[libmtj-java] 21/23: Imported Upstream version 1.0.1
Andreas Tille
tille at debian.org
Tue Sep 2 10:00:52 UTC 2014
This is an automated email from the git hooks/post-receive script.
tille pushed a commit to branch master
in repository libmtj-java.
commit 4566192a679771f5a969ea11363c30a2dc3e5779
Author: Andreas Tille <tille at debian.org>
Date: Tue Sep 2 11:56:37 2014 +0200
Imported Upstream version 1.0.1
---
.gitignore | 6 +
README.md | 123 ++
build.xml | 19 +
debian/changelog | 45 -
debian/compat | 1 -
debian/control | 54 -
debian/copyright | 62 -
debian/libmtj-java-doc.javadoc | 1 -
debian/libmtj-java.jlibs | 1 -
debian/libmtj-java.links | 1 -
debian/patches/series | 1 -
debian/patches/use_debian_jar_files.patch | 21 -
debian/rules | 10 -
debian/source/format | 1 -
debian/watch | 2 -
logging.properties | 12 +
pom.xml | 151 +++
.../no/uib/cipr/matrix/AbstractBandMatrix.java | 258 +++++
.../no/uib/cipr/matrix/AbstractDenseMatrix.java | 170 +++
.../java/no/uib/cipr/matrix/AbstractMatrix.java | 888 ++++++++++++++
.../no/uib/cipr/matrix/AbstractPackMatrix.java | 121 ++
.../no/uib/cipr/matrix/AbstractSymmBandMatrix.java | 168 +++
.../uib/cipr/matrix/AbstractSymmDenseMatrix.java | 291 +++++
.../no/uib/cipr/matrix/AbstractSymmPackMatrix.java | 180 +++
.../uib/cipr/matrix/AbstractTriangBandMatrix.java | 160 +++
.../uib/cipr/matrix/AbstractTriangDenseMatrix.java | 295 +++++
.../uib/cipr/matrix/AbstractTriangPackMatrix.java | 186 +++
.../java/no/uib/cipr/matrix/AbstractVector.java | 304 +++++
src/main/java/no/uib/cipr/matrix/BandCholesky.java | 254 ++++
src/main/java/no/uib/cipr/matrix/BandLU.java | 241 ++++
src/main/java/no/uib/cipr/matrix/BandMatrix.java | 255 ++++
.../java/no/uib/cipr/matrix/DenseCholesky.java | 235 ++++
src/main/java/no/uib/cipr/matrix/DenseLU.java | 203 ++++
src/main/java/no/uib/cipr/matrix/DenseMatrix.java | 517 +++++++++
src/main/java/no/uib/cipr/matrix/DenseVector.java | 328 ++++++
.../java/no/uib/cipr/matrix/DenseVectorSub.java | 45 +
src/main/java/no/uib/cipr/matrix/Diag.java | 39 +
src/main/java/no/uib/cipr/matrix/EVD.java | 201 ++++
.../java/no/uib/cipr/matrix/GivensRotation.java | 94 ++
src/main/java/no/uib/cipr/matrix/JobEig.java | 39 +
src/main/java/no/uib/cipr/matrix/JobEigRange.java | 47 +
src/main/java/no/uib/cipr/matrix/JobSVD.java | 62 +
src/main/java/no/uib/cipr/matrix/LQ.java | 138 +++
.../no/uib/cipr/matrix/LowerSPDBandMatrix.java | 84 ++
.../no/uib/cipr/matrix/LowerSPDDenseMatrix.java | 77 ++
.../no/uib/cipr/matrix/LowerSPDPackMatrix.java | 77 ++
.../no/uib/cipr/matrix/LowerSymmBandMatrix.java | 99 ++
.../no/uib/cipr/matrix/LowerSymmDenseMatrix.java | 99 ++
.../no/uib/cipr/matrix/LowerSymmPackMatrix.java | 106 ++
.../no/uib/cipr/matrix/LowerTriangBandMatrix.java | 110 ++
.../no/uib/cipr/matrix/LowerTriangDenseMatrix.java | 179 +++
.../no/uib/cipr/matrix/LowerTriangPackMatrix.java | 181 +++
src/main/java/no/uib/cipr/matrix/Matrices.java | 690 +++++++++++
src/main/java/no/uib/cipr/matrix/Matrix.java | 755 ++++++++++++
src/main/java/no/uib/cipr/matrix/MatrixEntry.java | 48 +
.../no/uib/cipr/matrix/MatrixNotSPDException.java | 47 +
.../uib/cipr/matrix/MatrixSingularException.java | 47 +
.../no/uib/cipr/matrix/NotConvergedException.java | 85 ++
.../no/uib/cipr/matrix/OrthogonalComputer.java | 103 ++
src/main/java/no/uib/cipr/matrix/PackCholesky.java | 233 ++++
.../java/no/uib/cipr/matrix/PermutationMatrix.java | 111 ++
src/main/java/no/uib/cipr/matrix/QL.java | 139 +++
src/main/java/no/uib/cipr/matrix/QR.java | 138 +++
src/main/java/no/uib/cipr/matrix/QRP.java | 224 ++++
src/main/java/no/uib/cipr/matrix/RQ.java | 138 +++
.../java/no/uib/cipr/matrix/SPDTridiagMatrix.java | 97 ++
src/main/java/no/uib/cipr/matrix/SVD.java | 215 ++++
src/main/java/no/uib/cipr/matrix/Side.java | 39 +
src/main/java/no/uib/cipr/matrix/SymmBandEVD.java | 177 +++
src/main/java/no/uib/cipr/matrix/SymmDenseEVD.java | 225 ++++
src/main/java/no/uib/cipr/matrix/SymmEVD.java | 100 ++
src/main/java/no/uib/cipr/matrix/SymmPackEVD.java | 174 +++
.../java/no/uib/cipr/matrix/SymmTridiagEVD.java | 180 +++
.../java/no/uib/cipr/matrix/SymmTridiagMatrix.java | 324 ++++++
src/main/java/no/uib/cipr/matrix/Transpose.java | 39 +
.../java/no/uib/cipr/matrix/TridiagMatrix.java | 293 +++++
.../uib/cipr/matrix/UnitLowerTriangBandMatrix.java | 113 ++
.../cipr/matrix/UnitLowerTriangDenseMatrix.java | 107 ++
.../uib/cipr/matrix/UnitLowerTriangPackMatrix.java | 100 ++
.../uib/cipr/matrix/UnitUpperTriangBandMatrix.java | 113 ++
.../cipr/matrix/UnitUpperTriangDenseMatrix.java | 109 ++
.../uib/cipr/matrix/UnitUpperTriangPackMatrix.java | 100 ++
src/main/java/no/uib/cipr/matrix/UpLo.java | 37 +
.../no/uib/cipr/matrix/UpperSPDBandMatrix.java | 84 ++
.../no/uib/cipr/matrix/UpperSPDDenseMatrix.java | 77 ++
.../no/uib/cipr/matrix/UpperSPDPackMatrix.java | 77 ++
.../no/uib/cipr/matrix/UpperSymmBandMatrix.java | 99 ++
.../no/uib/cipr/matrix/UpperSymmDenseMatrix.java | 99 ++
.../no/uib/cipr/matrix/UpperSymmPackMatrix.java | 106 ++
.../no/uib/cipr/matrix/UpperTriangBandMatrix.java | 109 ++
.../no/uib/cipr/matrix/UpperTriangDenseMatrix.java | 175 +++
.../no/uib/cipr/matrix/UpperTriangPackMatrix.java | 181 +++
src/main/java/no/uib/cipr/matrix/Vector.java | 185 +++
src/main/java/no/uib/cipr/matrix/VectorEntry.java | 43 +
.../java/no/uib/cipr/matrix/io/MatrixInfo.java | 276 +++++
.../java/no/uib/cipr/matrix/io/MatrixSize.java | 120 ++
.../no/uib/cipr/matrix/io/MatrixVectorReader.java | 658 +++++++++++
.../no/uib/cipr/matrix/io/MatrixVectorWriter.java | 531 +++++++++
.../java/no/uib/cipr/matrix/io/VectorInfo.java | 187 +++
.../java/no/uib/cipr/matrix/io/VectorSize.java | 84 ++
src/main/java/no/uib/cipr/matrix/io/package.html | 17 +
src/main/java/no/uib/cipr/matrix/overview.html | 11 +
src/main/java/no/uib/cipr/matrix/package.html | 131 +++
src/main/java/no/uib/cipr/matrix/sparse/AMG.java | 929 +++++++++++++++
.../matrix/sparse/AbstractIterationMonitor.java | 126 ++
.../matrix/sparse/AbstractIterativeSolver.java | 98 ++
.../java/no/uib/cipr/matrix/sparse/ArpackSym.java | 156 +++
.../java/no/uib/cipr/matrix/sparse/Arrays.java | 175 +++
src/main/java/no/uib/cipr/matrix/sparse/BiCG.java | 104 ++
.../java/no/uib/cipr/matrix/sparse/BiCGstab.java | 118 ++
src/main/java/no/uib/cipr/matrix/sparse/CG.java | 89 ++
src/main/java/no/uib/cipr/matrix/sparse/CGS.java | 107 ++
.../java/no/uib/cipr/matrix/sparse/Chebyshev.java | 121 ++
.../no/uib/cipr/matrix/sparse/CompColMatrix.java | 479 ++++++++
.../no/uib/cipr/matrix/sparse/CompDiagMatrix.java | 458 ++++++++
.../no/uib/cipr/matrix/sparse/CompRowMatrix.java | 561 +++++++++
.../matrix/sparse/DefaultIterationMonitor.java | 164 +++
.../cipr/matrix/sparse/DiagonalPreconditioner.java | 78 ++
.../uib/cipr/matrix/sparse/FlexCompColMatrix.java | 253 ++++
.../uib/cipr/matrix/sparse/FlexCompRowMatrix.java | 267 +++++
src/main/java/no/uib/cipr/matrix/sparse/GMRES.java | 178 +++
src/main/java/no/uib/cipr/matrix/sparse/ICC.java | 155 +++
src/main/java/no/uib/cipr/matrix/sparse/ILU.java | 141 +++
src/main/java/no/uib/cipr/matrix/sparse/ILUT.java | 446 +++++++
src/main/java/no/uib/cipr/matrix/sparse/IR.java | 72 ++
.../no/uib/cipr/matrix/sparse/ISparseVector.java | 24 +
.../uib/cipr/matrix/sparse/IterationMonitor.java | 119 ++
.../uib/cipr/matrix/sparse/IterationReporter.java | 52 +
.../no/uib/cipr/matrix/sparse/IterativeSolver.java | 76 ++
.../IterativeSolverNotConvergedException.java | 91 ++
.../uib/cipr/matrix/sparse/LinkedSparseMatrix.java | 457 ++++++++
.../cipr/matrix/sparse/MatrixIterationMonitor.java | 131 +++
.../cipr/matrix/sparse/NoIterationReporter.java | 38 +
.../matrix/sparse/OutputIterationReporter.java | 64 +
.../no/uib/cipr/matrix/sparse/Preconditioner.java | 65 ++
src/main/java/no/uib/cipr/matrix/sparse/QMR.java | 222 ++++
src/main/java/no/uib/cipr/matrix/sparse/SSOR.java | 201 ++++
.../no/uib/cipr/matrix/sparse/SparseVector.java | 442 +++++++
.../no/uib/cipr/matrix/sparse/SuperIterator.java | 158 +++
.../cipr/matrix/sparse/UnitLowerCompRowMatrix.java | 88 ++
.../uib/cipr/matrix/sparse/UpperCompRowMatrix.java | 92 ++
.../java/no/uib/cipr/matrix/sparse/package.html | 63 +
.../cipr/matrix/sparse/LinkedSparseMatrixPerf.R | 111 ++
.../java/no/uib/cipr/matrix/BandCholeskyTest.java | 130 +++
src/test/java/no/uib/cipr/matrix/BandLUTest.java | 110 ++
.../java/no/uib/cipr/matrix/BandMatrixTest.java | 60 +
.../java/no/uib/cipr/matrix/DenseCholeskyTest.java | 126 ++
src/test/java/no/uib/cipr/matrix/DenseLUTest.java | 120 ++
.../java/no/uib/cipr/matrix/DenseMatrixTest.java | 123 ++
.../no/uib/cipr/matrix/DenseVectorSubTest.java | 23 +
.../java/no/uib/cipr/matrix/DenseVectorTest.java | 41 +
src/test/java/no/uib/cipr/matrix/LQTest.java | 84 ++
.../no/uib/cipr/matrix/LowerSPDBandMatrixTest.java | 43 +
.../uib/cipr/matrix/LowerSPDDenseMatrixTest.java | 42 +
.../no/uib/cipr/matrix/LowerSPDPackMatrixTest.java | 42 +
.../uib/cipr/matrix/LowerSymmBandMatrixTest.java | 43 +
.../uib/cipr/matrix/LowerSymmDenseMatrixTest.java | 42 +
.../uib/cipr/matrix/LowerSymmPackMatrixTest.java | 42 +
.../uib/cipr/matrix/LowerTriangBandMatrixTest.java | 45 +
.../cipr/matrix/LowerTriangDenseMatrixTest.java | 44 +
.../uib/cipr/matrix/LowerTriangPackMatrixTest.java | 44 +
.../no/uib/cipr/matrix/MatrixTestAbstract.java | 1224 ++++++++++++++++++++
.../no/uib/cipr/matrix/OrthogonalTestAbstract.java | 66 ++
.../java/no/uib/cipr/matrix/PackCholeskyTest.java | 125 ++
.../no/uib/cipr/matrix/PermutationMatrixTest.java | 64 +
src/test/java/no/uib/cipr/matrix/QLTest.java | 84 ++
src/test/java/no/uib/cipr/matrix/QRPTest.java | 181 +++
src/test/java/no/uib/cipr/matrix/QRTest.java | 84 ++
src/test/java/no/uib/cipr/matrix/RQTest.java | 84 ++
.../no/uib/cipr/matrix/SPDTridiagMatrixTest.java | 42 +
.../java/no/uib/cipr/matrix/SingularvalueTest.java | 85 ++
.../no/uib/cipr/matrix/SquareDenseMatrixTest.java | 42 +
.../matrix/StructImmutableMatrixTestAbstract.java | 142 +++
.../no/uib/cipr/matrix/SymmBandEigenvalueTest.java | 94 ++
.../uib/cipr/matrix/SymmDenseEigenvalueTest.java | 93 ++
.../cipr/matrix/SymmEigenvalueTestAbstract.java | 74 ++
.../no/uib/cipr/matrix/SymmPackEigenvalueTest.java | 93 ++
.../uib/cipr/matrix/SymmTridiagEigenvalueTest.java | 67 ++
.../no/uib/cipr/matrix/SymmTridiagMatrixTest.java | 42 +
.../cipr/matrix/SymmetricMatrixTestAbstract.java | 79 ++
.../uib/cipr/matrix/TriangMatrixTestAbstract.java | 37 +
.../java/no/uib/cipr/matrix/TridiagMatrixTest.java | 51 +
.../cipr/matrix/UnitLowerTriangBandMatrixTest.java | 43 +
.../matrix/UnitLowerTriangDenseMatrixTest.java | 59 +
.../cipr/matrix/UnitLowerTriangPackMatrixTest.java | 42 +
.../cipr/matrix/UnitTriangMatrixTestAbstract.java | 96 ++
.../cipr/matrix/UnitUpperTriangBandMatrixTest.java | 43 +
.../matrix/UnitUpperTriangDenseMatrixTest.java | 42 +
.../cipr/matrix/UnitUpperTriangPackMatrixTest.java | 42 +
.../no/uib/cipr/matrix/UpperSPDBandMatrixTest.java | 43 +
.../uib/cipr/matrix/UpperSPDDenseMatrixTest.java | 42 +
.../no/uib/cipr/matrix/UpperSPDPackMatrixTest.java | 42 +
.../uib/cipr/matrix/UpperSymmBandMatrixTest.java | 43 +
.../uib/cipr/matrix/UpperSymmDenseMatrixTest.java | 42 +
.../uib/cipr/matrix/UpperSymmPackMatrixTest.java | 42 +
.../uib/cipr/matrix/UpperTriangBandMatrixTest.java | 45 +
.../cipr/matrix/UpperTriangDenseMatrixTest.java | 44 +
.../uib/cipr/matrix/UpperTriangPackMatrixTest.java | 44 +
src/test/java/no/uib/cipr/matrix/Utilities.java | 516 +++++++++
.../no/uib/cipr/matrix/VectorTestAbstract.java | 296 +++++
.../no/uib/cipr/matrix/io/MatrixVectorIoTest.java | 28 +
.../no/uib/cipr/matrix/sparse/ArpackSymTest.java | 76 ++
.../uib/cipr/matrix/sparse/BiCGDiagonalTest.java | 40 +
.../no/uib/cipr/matrix/sparse/BiCGILUTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/BiCGTest.java | 39 +
.../cipr/matrix/sparse/BiCGstabDiagonalTest.java | 40 +
.../uib/cipr/matrix/sparse/BiCGstabILUTTest.java | 41 +
.../no/uib/cipr/matrix/sparse/BiCGstabILUTest.java | 41 +
.../no/uib/cipr/matrix/sparse/BiCGstabTest.java | 39 +
.../java/no/uib/cipr/matrix/sparse/CGAMGTest.java | 40 +
.../no/uib/cipr/matrix/sparse/CGDiagonalTest.java | 40 +
.../java/no/uib/cipr/matrix/sparse/CGICCTest.java | 41 +
.../no/uib/cipr/matrix/sparse/CGSDiagonalTest.java | 40 +
.../no/uib/cipr/matrix/sparse/CGSILUTTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/CGSILUTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/CGSSORTest.java | 42 +
.../java/no/uib/cipr/matrix/sparse/CGSTest.java | 39 +
.../java/no/uib/cipr/matrix/sparse/CGTest.java | 39 +
.../uib/cipr/matrix/sparse/ChebyshevAMGTest.java | 40 +
.../cipr/matrix/sparse/ChebyshevDiagonalTest.java | 40 +
.../uib/cipr/matrix/sparse/ChebyshevICCTest.java | 41 +
.../uib/cipr/matrix/sparse/ChebyshevSSORTest.java | 42 +
.../no/uib/cipr/matrix/sparse/ChebyshevTest.java | 50 +
.../uib/cipr/matrix/sparse/CompColMatrixTest.java | 45 +
.../uib/cipr/matrix/sparse/CompDiagMatrixTest.java | 43 +
.../uib/cipr/matrix/sparse/CompRowMatrixTest.java | 45 +
.../cipr/matrix/sparse/FlexCompColMatrixTest.java | 44 +
.../cipr/matrix/sparse/FlexCompRowMatrixTest.java | 44 +
.../uib/cipr/matrix/sparse/GMRESDiagonalTest.java | 40 +
.../no/uib/cipr/matrix/sparse/GMRESILUTTest.java | 41 +
.../no/uib/cipr/matrix/sparse/GMRESILUTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/GMRESTest.java | 39 +
.../java/no/uib/cipr/matrix/sparse/ICCTest.java | 46 +
.../java/no/uib/cipr/matrix/sparse/ILUTTest.java | 46 +
.../java/no/uib/cipr/matrix/sparse/ILUTest.java | 45 +
.../IncompleteFactorizationTestAbstract.java | 79 ++
.../matrix/sparse/IterativeSolverTestAbstract.java | 178 +++
.../cipr/matrix/sparse/LinkedSparseMatrixTest.java | 214 ++++
.../no/uib/cipr/matrix/sparse/QMRDiagonalTest.java | 40 +
.../no/uib/cipr/matrix/sparse/QMRILUTTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/QMRILUTest.java | 41 +
.../java/no/uib/cipr/matrix/sparse/QMRTest.java | 39 +
.../sparse/SPDIterativeSolverTestAbstract.java | 53 +
.../matrix/sparse/SparseMatrixTestAbstract.java | 56 +
.../SparseStructImmutableMatrixTestAbstract.java | 57 +
.../uib/cipr/matrix/sparse/SparseVectorTest.java | 100 ++
246 files changed, 31362 insertions(+), 200 deletions(-)
diff --git a/.gitignore b/.gitignore
new file mode 100644
index 0000000..0f182a0
--- /dev/null
+++ b/.gitignore
@@ -0,0 +1,6 @@
+*.class
+
+# Package Files #
+*.jar
+*.war
+*.ear
diff --git a/README.md b/README.md
new file mode 100644
index 0000000..9159cd5
--- /dev/null
+++ b/README.md
@@ -0,0 +1,123 @@
+matrix-toolkits-java
+====================
+
+**MTJ** is a high-performance library for developing linear algebra applications.
+
+MTJ is based on [BLAS](http://www.netlib.org/blas) and [LAPACK](http://www.netlib.org/lapack) for its dense and structured sparse computations, and on the [Templates](http://www.netlib.org/templates) project for unstructured sparse operations.
+
+MTJ uses the [`netlib-java`](https://github.com/fommil/netlib-java/) project as a backend,
+which will automatically use machine-optimised natives, if they are available. Please read the [`netlib-java` documentation](https://github.com/fommil/netlib-java/) for the extra steps needed to ensure that you are getting the best performance for your system.
+
+Performance to Other Libraries
+==============================
+
+*I am currently running the [java-matrix-benchmark](https://github.com/fommil/matrix-toolkits-java/issues/33). Results will be posted here when they are ready... they take a week!!*
+
+
+Sparse Storage
+==============
+
+A variety of sparse matrix / vector storage classes are available:
+
+* [`CompColMatrix`](src/main/java/no/uib/cipr/matrix/sparse/CompColMatrix.java)
+* [`CompDiagMatrix`](src/main/java/no/uib/cipr/matrix/sparse/CompDiagMatrix.java)
+* [`CompRowMatrix`](src/main/java/no/uib/cipr/matrix/sparse/CompRowMatrix.java)
+* [`FlexCompColMatrix`](src/main/java/no/uib/cipr/matrix/sparse/FlexCompColMatrix.java)
+* [`FlexCompRowMatrix`](src/main/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrix.java)
+* [`UnitLowerCompRowMatrix`](src/main/java/no/uib/cipr/matrix/sparse/UnitLowerCompRowMatrix.java)
+* [`UpperCompRowMatrix`](src/main/java/no/uib/cipr/matrix/sparse/UpperCompRowMatrix.java)
+* [`SparseVector`](src/main/java/no/uib/cipr/matrix/sparse/SparseVector.java)
+* [`LinkedSparseMatrix`](src/main/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrix.java)
+
+The `LinkedSparseMatrix` storage type is a novel storage type developed under this project. It maintains two tail links, one for the next matrix element by row order and another by column order. Lookups are kept into each row and column, making multiplication and transpose multiplication very fast.
+
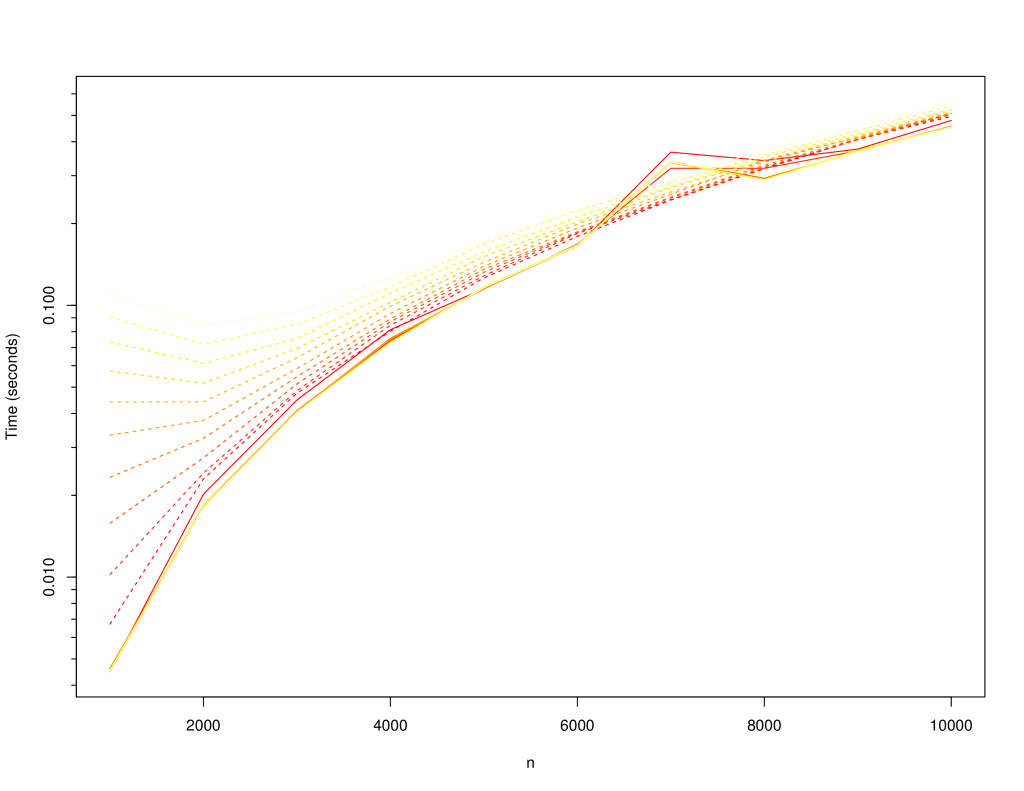
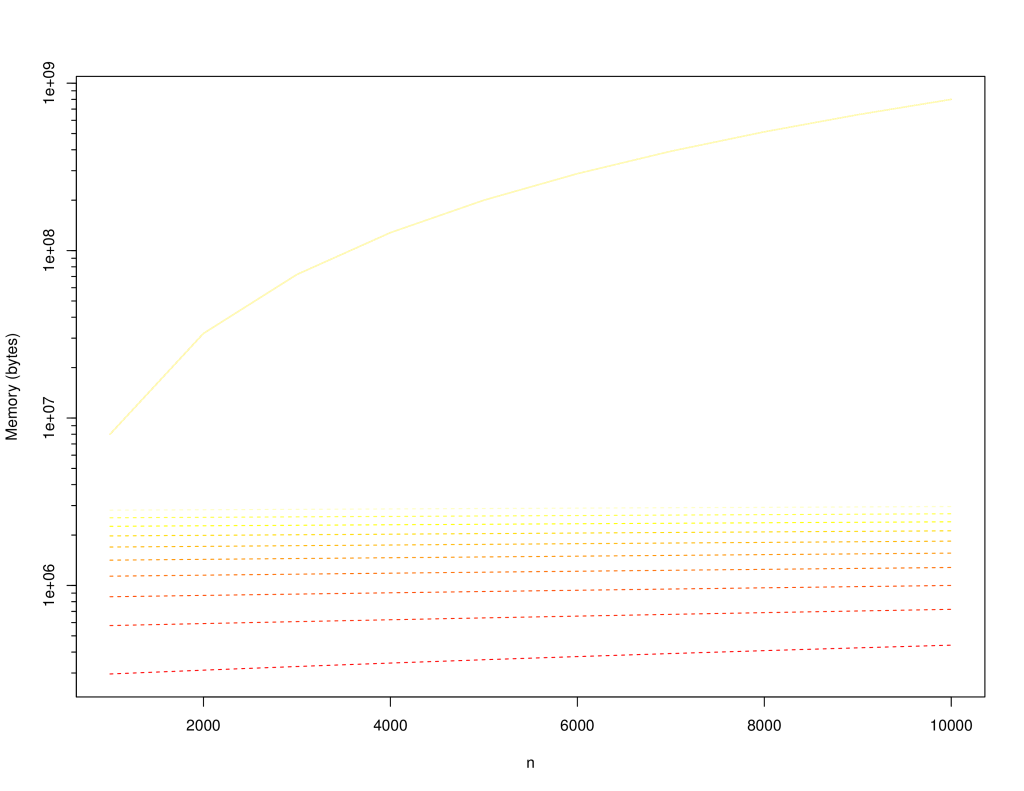
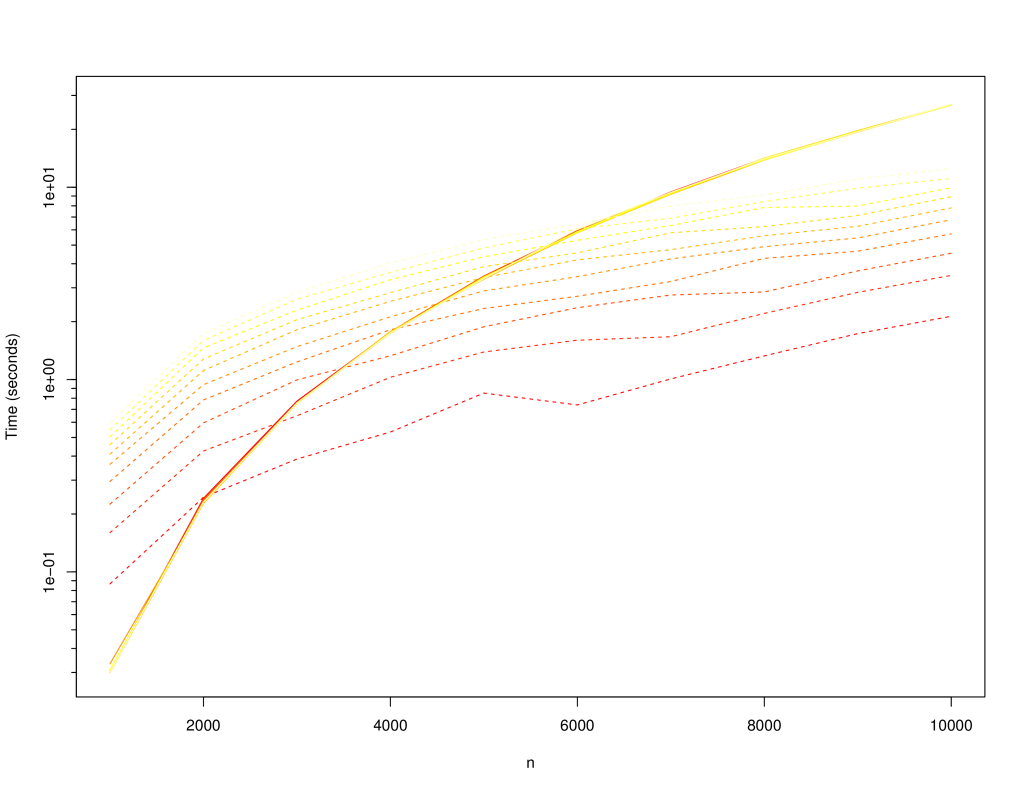
+The following charts compare the `LinkedSparseMatrix` against `DenseMatrix` for increasing matrix size (`n x n`) and number of non-zero elements, `m`. Lighter lines indicate larger `m`: varied from `10,000` to `100,000`. Solid lines are for dense matrix, dashed lines are the sparse matrix.
+
+The following is time to initialise the matrix:
+
+
+
+The following is the memory consumption:
+
+
+
+The following is the time to perform a multiplication with a dense matrix and output into a dense matrix:
+
+
+
+
+Sparse Solvers
+==============
+
+MTJ provides [ARPACK](http://www.caam.rice.edu/software/ARPACK/) for very large symmetric matrices in [ArpackSym](src/main/java/no/uib/cipr/matrix/sparse/ArpackSym.java) (see the example usage in [ArpackSymTest](src/test/java/no/uib/cipr/matrix/sparse/ArpackSymTest.java)). ARPACK solves an arbitrary number of eigenvalues / eigenvectors.
+
+In addition, implementations of the netlib Templates are available in the [`no.uib.cipr.matrix.sparse`](src/test/java/no/uib/cipr/matrix/sparse) package.
+
+Users may wish to look at [Sparse Eigensolvers for Java](http://code.google.com/p/sparse-eigensolvers-java/) for another solver.
+
+
+Licence
+=======
+
+Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+Copyright (C) 2006-2013 Samuel Halliday
+
+This program is free software: you can redistribute it and/or modify
+it under the terms of the GNU Lesser General Public License as published by
+the Free Software Foundation, either version 3 of the License, or
+(at your option) any later version.
+
+This program is distributed in the hope that it will be useful,
+but WITHOUT ANY WARRANTY; without even the implied warranty of
+MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
+GNU Lesser General Public License for more details.
+
+You should have received a copy of the GNU Lesser General Public License
+along with this program. If not, see http://www.gnu.org/licenses/
+
+History
+=======
+
+This project was originally written by Bjørn-Ove Heimsund, who has taken a step back due to other commitments. The group here is primarily concerned with keeping the library maintained, and fixing bugs as they are discovered. There is no road plan for future releases.
+
+Installation
+============
+
+Releases are distributed on Maven central:
+
+```xml
+<dependency>
+ <groupId>com.googlecode.matrix-toolkits-java</groupId>
+ <artifactId>mtj</artifactId>
+ <version>1.0</version>
+</dependency>
+```
+
+Unofficial single-jar builds may be available from [`java-matrix-benchmark`](https://code.google.com/p/java-matrix-benchmark/source/browse/#svn%2Ftrunk%2Flib%2Fmtj) for laggards who don't have [5 minutes to learn Maven](http://maven.apache.org/guides/getting-started/maven-in-five-minutes.html).
+
+
+Snapshots are distributed on Sonatype's Snapshot Repository:
+
+```xml
+<dependency>
+ <groupId>com.googlecode.matrix-toolkits-java</groupId>
+ <artifactId>mtj</artifactId>
+ <version>1.1-SNAPSHOT</version>
+</dependency>
+```
+
+Donations
+=========
+
+Please consider supporting the maintenance of this open source project by starring it, above, and with a donation:
+
+[](https://www.paypal.com/cgi-bin/webscr?cmd=_donations&business=B2HW5ATB8C3QW&lc=GB&item_name=mtj¤cy_code=GBP&bn=PP%2dDonationsBF%3abtn_donateCC_LG%2egif%3aNonHosted)
+
+
+Contributing
+============
+
+Contributors are encouraged to fork this repository and issue pull
+requests. Contributors implicitly agree to assign an unrestricted licence
+to Sam Halliday, but retain the copyright of their code (this means
+we both have the freedom to update the licence for those contributions).
diff --git a/build.xml b/build.xml
new file mode 100644
index 0000000..e5d049d
--- /dev/null
+++ b/build.xml
@@ -0,0 +1,19 @@
+<?xml version="1.0" encoding="UTF-8"?>
+<project name="Matrix_Toolkits_for_Java" default="default" basedir=".">
+ <description>Builds, tests, and runs the project Matrix Toolkits for Java.</description>
+ <property name="version" value="dev" />
+ <import file="nbproject/build-impl.xml"/>
+
+ <target name="-post-clean">
+ <delete dir="javadoc" />
+ </target>
+
+ <target name="package" depends="jar, javadoc">
+ <!-- TODO: consider bundling BLAS/LAPACK -->
+ <move file="dist/javadoc" todir="."/>
+ <move file="${dist.jar}" tofile="mtj-${version}.jar" />
+ <zip destfile="mtj-${version}.zip">
+ <zipfileset dir="." excludes="**/lib/nblibraries-private.properties **/nbproject/private** **/.ant-targets-build.xml dist/** build/** **/*.zip **/.svn**" prefix="mtj-${version}" />
+ </zip>
+ </target>
+</project>
diff --git a/debian/changelog b/debian/changelog
deleted file mode 100644
index 22a2f8f..0000000
--- a/debian/changelog
+++ /dev/null
@@ -1,45 +0,0 @@
-mtj (1.0.1-1) UNRELEASED; urgency=medium
-
- * New upstream version
- * d/watch: moved to Github
- * d/get-orig-source: deleted since no need to repack any more
- * Move from Debian Med team to pkg-java team maintenance
-
- -- Andreas Tille <tille at debian.org> Thu, 28 Aug 2014 14:31:28 +0200
-
-mtj (0.9.14+dfsg-2) unstable; urgency=low
-
- * Upload to unstable
- Closes: #720567
- * debian/control:
- - cme fix dpkg-control
- - anonscm in Vcs fields
-
- -- Andreas Tille <tille at debian.org> Fri, 23 Aug 2013 14:40:06 +0200
-
-mtj (0.9.14+dfsg-1) experimental; urgency=low
-
- * debian/copyright:
- - DEP5
- - Add Files-Excluded to document what was removed from original source
- * debian/control: Standards-Version: 3.9.4 (no changes needed)
- * debian/{control,rules}: use default java implementation
- (thanks for the patch to James Page <james.page at ubuntu.com>)
- Closes: #684160
-
- -- Andreas Tille <tille at debian.org> Mon, 17 Dec 2012 14:28:38 +0100
-
-mtj (0.9.14~dfsg-2) unstable; urgency=low
-
- * debian/control: package should not be in main because it
- (unfortunately) (build-)depends from another package in
- contrib (libnetlib-java)
- Closes: #676766
-
- -- Andreas Tille <tille at debian.org> Sun, 10 Jun 2012 09:20:13 +0200
-
-mtj (0.9.14~dfsg-1) unstable; urgency=low
-
- * Initial release (Closes: #675919)
-
- -- Andreas Tille <tille at debian.org> Sun, 03 Jun 2012 13:02:47 +0200
diff --git a/debian/compat b/debian/compat
deleted file mode 100644
index ec63514..0000000
--- a/debian/compat
+++ /dev/null
@@ -1 +0,0 @@
-9
diff --git a/debian/control b/debian/control
deleted file mode 100644
index 3a82c01..0000000
--- a/debian/control
+++ /dev/null
@@ -1,54 +0,0 @@
-Source: mtj
-Maintainer: Debian Java Maintainers <pkg-java-maintainers at lists.alioth.debian.org>
-Uploaders: Andreas Tille <tille at debian.org>
-Section: contrib/java
-Priority: optional
-Build-Depends: debhelper (>= 9),
- javahelper,
- default-jdk (>= 1:1.6),
- ant,
- libnetlib-java
-Standards-Version: 3.9.4
-Vcs-Git: git://anonscm.debian.org/pkg-java/libmtj-java.git
-Vcs-Browser: http://anonscm.debian.org/gitweb/?p=pkg-java/libmtj-java.git
-Homepage: http://code.google.com/p/matrix-toolkits-java/
-
-Package: libmtj-java
-Architecture: all
-Depends: ${misc:Depends},
- ${java:Depends}
-Recommends: ${java:Recommends}
-Description: Java library for developing numerical applications
- MTJ is designed to be used as a library for developing numerical
- applications, both for small and large scale computations. The library
- is based on BLAS and LAPACK for its dense and structured sparse
- computations, and on the Templates project for unstructured sparse
- operations.
- .
- MTJ uses the netlib-java project as a backend, which can be set up to
- use machine-optimised BLAS libraries for improved performance of dense
- matrix operations, falling back to a pure Java implementation. This
- ensures perfect portability, while allowing for improved performance in
- a production environment.
-
-Package: libmtj-java-doc
-Architecture: all
-Section: contrib/doc
-Depends: ${misc:Depends},
- ${java:Depends}
-Recommends: ${java:Recommends}
-Description: Java library for developing numerical applications (documentation)
- MTJ is designed to be used as a library for developing numerical
- applications, both for small and large scale computations. The library
- is based on BLAS and LAPACK for its dense and structured sparse
- computations, and on the Templates project for unstructured sparse
- operations.
- .
- MTJ uses the netlib-java project as a backend, which can be set up to
- use machine-optimised BLAS libraries for improved performance of dense
- matrix operations, falling back to a pure Java implementation. This
- ensures perfect portability, while allowing for improved performance in
- a production environment.
- .
- This package contains the javadoc documentation files.
-
diff --git a/debian/copyright b/debian/copyright
deleted file mode 100644
index be0b6f5..0000000
--- a/debian/copyright
+++ /dev/null
@@ -1,62 +0,0 @@
-Format: http://www.debian.org/doc/packaging-manuals/copyright-format/1.0/
-Upstream-Name: matrix-toolkits-java
-Upstream-Contact: Sam Halliday <Sam.Halliday at gmail.com>
- Bjørn-Ove Heimsund <Bjorn.Ove.Heimsund at gmail.com>
-Source: http://code.google.com/p/matrix-toolkits-java/downloads/list
-
-Files: *
-Copyright: 2008-2011 Sam Halliday <Sam.Halliday at gmail.com>
- Bjørn-Ove Heimsund <Bjorn.Ove.Heimsund at gmail.com>
-License: LGPL-3+
-
-Files: debian/*
-Copyright: 2012 Andreas Tille <tille at debian.org>
- Olivier Sallou <osallou at debian.org>
-License: LGPL-3+
-
-Files: lib/netlib-java/*.jar
-Copyright: 2008-2011 Sam Halliday <Sam.Halliday at gmail.com>
- Keith Seymour <keithseymour at gmail.com>
-License: BSD-new
- Redistribution and use in source and binary forms, with or without
- modification, are permitted provided that the following conditions are
- met:
- .
- * Redistributions of source code must retain the above copyright
- notice, this list of conditions and the following disclaimer.
- * Redistributions in binary form must reproduce the above copyright
- notice, this list of conditions and the following disclaimer in the
- documentation and/or other materials provided with the distribution.
- .
- THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS
- IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED
- TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A
- PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT
- HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL,
- SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED
- TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR
- PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF
- LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING
- NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
- SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
-Comment: The files are provided as binary JAR files. The source is
- available from
- http://code.google.com/p/netlib-java/
-
-License: LGPL-3+
- This program is free software: you can redistribute it and/or modify
- it under the terms of the GNU Lesser General Public License as published by
- the Free Software Foundation, either version 3 of the License, or
- (at your option) any later version.
- .
- This program is distributed in the hope that it will be useful,
- but WITHOUT ANY WARRANTY; without even the implied warranty of
- MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
- GNU General Public License for more details.
- .
- You should have received a copy of the GNU Lesser General Public License
- along with this program. If not, see <http://www.gnu.org/licenses/>.
- .
- On a Debian GNU/Linux system, the GNU Lesser GPL license version 3 is included
- in the file ‘/usr/share/common-licenses/LGPL-3’, and the GNU GPL license
- version 3 is included in the file ‘/usr/share/common-licenses/GPL-3’.
diff --git a/debian/libmtj-java-doc.javadoc b/debian/libmtj-java-doc.javadoc
deleted file mode 100644
index af30b98..0000000
--- a/debian/libmtj-java-doc.javadoc
+++ /dev/null
@@ -1 +0,0 @@
-dist/javadoc /usr/share/doc/libmtj-java
diff --git a/debian/libmtj-java.jlibs b/debian/libmtj-java.jlibs
deleted file mode 100644
index 77731a9..0000000
--- a/debian/libmtj-java.jlibs
+++ /dev/null
@@ -1 +0,0 @@
-dist/Matrix_Toolkits_for_Java.jar
diff --git a/debian/libmtj-java.links b/debian/libmtj-java.links
deleted file mode 100644
index c5bd338..0000000
--- a/debian/libmtj-java.links
+++ /dev/null
@@ -1 +0,0 @@
-usr/share/java/Matrix_Toolkits_for_Java.jar usr/share/java/mtj.jar
diff --git a/debian/patches/series b/debian/patches/series
deleted file mode 100644
index 61a6094..0000000
--- a/debian/patches/series
+++ /dev/null
@@ -1 +0,0 @@
-use_debian_jar_files.patch
diff --git a/debian/patches/use_debian_jar_files.patch b/debian/patches/use_debian_jar_files.patch
deleted file mode 100644
index a31a994..0000000
--- a/debian/patches/use_debian_jar_files.patch
+++ /dev/null
@@ -1,21 +0,0 @@
---- mtj-0.9.14.orig/build.xml
-+++ mtj-0.9.14/build.xml
-@@ -16,4 +16,6 @@
- <zipfileset dir="." excludes="**/lib/nblibraries-private.properties **/nbproject/private** **/.ant-targets-build.xml dist/** build/** **/*.zip **/.svn**" prefix="mtj-${version}" />
- </zip>
- </target>
-+ <property name="src.dir" value="src"/>
-+ <property name="javac.classpath" value="/usr/share/java/junit.jar:/usr/share/java/f2jutil.jar:/usr/share/java/netlib-java.jar"/>
- </project>
---- mtj-0.9.14.orig/lib/nblibraries.properties
-+++ mtj-0.9.14/lib/nblibraries.properties
-@@ -1,7 +1,6 @@
- libs.junit.classpath=\
-- ${base}/junit/junit-3.8.2.jar
-+ /usr/share/java/junit.jar
- libs.junit.javadoc=\
- ${base}/junit/junit-3.8.2-api.zip
- libs.netlib-java.classpath=\
-- ${base}/netlib-java/arpack_combo-0.1.jar:\
-- ${base}/netlib-java/netlib-java-0.9.3.jar
-+ /usr/share/java/netlib-java.jar
diff --git a/debian/rules b/debian/rules
deleted file mode 100755
index 0ee2477..0000000
--- a/debian/rules
+++ /dev/null
@@ -1,10 +0,0 @@
-#!/usr/bin/make -f
-
-JAVA_HOME=/usr/lib/jvm/default-java
-
-%:
- dh $@ --with javahelper
-
-override_dh_auto_build:
- dh_auto_build -- -Dant.build.javac.source=1.5 -Dant.build.javac.target=1.5
-
diff --git a/debian/source/format b/debian/source/format
deleted file mode 100644
index 163aaf8..0000000
--- a/debian/source/format
+++ /dev/null
@@ -1 +0,0 @@
-3.0 (quilt)
diff --git a/debian/watch b/debian/watch
deleted file mode 100644
index 632dd59..0000000
--- a/debian/watch
+++ /dev/null
@@ -1,2 +0,0 @@
-version=3
-https://github.com/fommil/matrix-toolkits-java/releases .*/archive/mtj-(\d[\d.-]+)\.(?:tar(?:\.gz|\.bz2)?|tgz)
diff --git a/logging.properties b/logging.properties
new file mode 100644
index 0000000..5d158da
--- /dev/null
+++ b/logging.properties
@@ -0,0 +1,12 @@
+# -Djava.util.logging.config.file=logging.properties
+handlers = java.util.logging.ConsoleHandler
+
+java.util.logging.ConsoleHandler.level = ALL
+java.util.logging.ConsoleHandler.formatter = com.github.fommil.logging.CustomFormatter
+
+com.github.fommil.logging.CustomFormatter.format = %L: %m [%c] %e %E%S
+com.github.fommil.logging.CustomFormatter.stackExclude = org.jetbrains. com.intellij java. sun. com.sun
+
+.level = WARNING
+com.github.fommil.level = INFO
+no.uib.cipr.matrix.sparse.level = INFO
diff --git a/pom.xml b/pom.xml
new file mode 100644
index 0000000..5466d4a
--- /dev/null
+++ b/pom.xml
@@ -0,0 +1,151 @@
+<?xml version="1.0" encoding="UTF-8"?>
+<project xmlns="http://maven.apache.org/POM/4.0.0" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://maven.apache.org/POM/4.0.0 http://maven.apache.org/xsd/maven-4.0.0.xsd">
+ <modelVersion>4.0.0</modelVersion>
+ <groupId>com.googlecode.matrix-toolkits-java</groupId>
+ <artifactId>mtj</artifactId>
+ <version>1.0.1</version>
+ <packaging>jar</packaging>
+ <properties>
+ <project.build.sourceEncoding>UTF-8</project.build.sourceEncoding>
+ </properties>
+ <prerequisites>
+ <maven>3.0.3</maven>
+ </prerequisites>
+ <parent>
+ <groupId>org.sonatype.oss</groupId>
+ <artifactId>oss-parent</artifactId>
+ <version>7</version>
+ </parent>
+ <name>Matrix Toolkits for Java</name>
+ <description>A comprehensive collection of matrix data structures, linear solvers, least squares methods,
+ eigenvalue, and singular value decompositions.
+ </description>
+ <url>https://github.com/fommil/matrix-toolkits-java/</url>
+ <licenses>
+ <license>
+ <name>GNU Lesser General Public License</name>
+ <url>http://www.gnu.org/licenses/lgpl.html</url>
+ </license>
+ </licenses>
+ <scm>
+ <url>https://github.com/fommil/matrix-toolkits-java</url>
+ <connection>scm:git:git at github.com:fommil/matrix-toolkits-java.git</connection>
+ <developerConnection>scm:git:git at github.com:fommil/matrix-toolkits-java.git</developerConnection>
+ </scm>
+ <developers>
+ <developer>
+ <name>Bjørn-Ove Heimsund</name>
+ </developer>
+ <developer>
+ <name>Sam Halliday</name>
+ <id>fommil</id>
+ <email>sam.halliday at gmail.com</email>
+ </developer>
+ </developers>
+ <issueManagement>
+ <system>github</system>
+ <url>https://github.com/fommil/matrix-toolkits-java/issues</url>
+ </issueManagement>
+ <repositories>
+ <repository>
+ <id>sonatype-snapshots</id>
+ <url>https://oss.sonatype.org/content/repositories/snapshots/</url>
+ <releases>
+ <enabled>false</enabled>
+ </releases>
+ <snapshots>
+ <enabled>true</enabled>
+ </snapshots>
+ </repository>
+ <repository>
+ <id>sonatype-releases</id>
+ <url>https://oss.sonatype.org/content/repositories/releases/</url>
+ <releases>
+ <enabled>true</enabled>
+ </releases>
+ <snapshots>
+ <enabled>false</enabled>
+ </snapshots>
+ </repository>
+ </repositories>
+ <!-- mvn versions:display-dependency-updates -->
+ <dependencies>
+ <dependency>
+ <groupId>com.github.fommil.netlib</groupId>
+ <artifactId>all</artifactId>
+ <version>1.1.1</version>
+ <type>pom</type>
+ </dependency>
+ <dependency>
+ <groupId>org.projectlombok</groupId>
+ <artifactId>lombok</artifactId>
+ <version>1.12.2</version>
+ <scope>provided</scope>
+ </dependency>
+ <dependency>
+ <groupId>com.github.fommil</groupId>
+ <artifactId>java-logging</artifactId>
+ <version>1.1</version>
+ <scope>provided</scope>
+ </dependency>
+ <dependency>
+ <groupId>junit</groupId>
+ <artifactId>junit</artifactId>
+ <version>3.8.2</version>
+ <scope>test</scope>
+ </dependency>
+ <dependency>
+ <groupId>com.google.guava</groupId>
+ <artifactId>guava</artifactId>
+ <version>15.0</version>
+ <scope>test</scope>
+ </dependency>
+ <dependency>
+ <groupId>net.sf.opencsv</groupId>
+ <artifactId>opencsv</artifactId>
+ <version>2.3</version>
+ <scope>test</scope>
+ </dependency>
+ </dependencies>
+ <!-- mvn versions:display-plugin-updates -->
+ <build>
+ <pluginManagement>
+ <plugins>
+ <plugin>
+ <groupId>org.apache.maven.plugins</groupId>
+ <artifactId>maven-gpg-plugin</artifactId>
+ <version>1.4</version>
+ <executions>
+ <execution>
+ <id>sign-artifacts</id>
+ <phase>verify</phase>
+ <goals>
+ <goal>sign</goal>
+ </goals>
+ </execution>
+ </executions>
+ </plugin>
+ <plugin>
+ <groupId>org.apache.maven.plugins</groupId>
+ <artifactId>maven-compiler-plugin</artifactId>
+ <version>3.1</version>
+ <configuration>
+ <source>1.6</source>
+ <target>1.6</target>
+ </configuration>
+ </plugin>
+ <plugin>
+ <!-- For the benefit of java-matrix-benchmark -->
+ <!-- mvn compile assembly:single -->
+ <artifactId>maven-assembly-plugin</artifactId>
+ <version>2.4</version>
+ <configuration>
+ <descriptorRefs>
+ <descriptorRef>jar-with-dependencies</descriptorRef>
+ </descriptorRefs>
+ </configuration>
+ </plugin>
+ </plugins>
+ </pluginManagement>
+ </build>
+</project>
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractBandMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractBandMatrix.java
new file mode 100644
index 0000000..9bcbbca
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractBandMatrix.java
@@ -0,0 +1,258 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+/**
+ * Partial implementation of a banded matrix
+ */
+abstract class AbstractBandMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix data
+ */
+ double[] data;
+
+ /**
+ * Number of upper and lower diagonals
+ */
+ int kl, ku;
+
+ /**
+ * Size of the matrix. It is always square
+ */
+ int n;
+
+ /**
+ * Constructor for AbstractBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kl
+ * Number of diagonals below the main diagonal
+ * @param ku
+ * Number of diagonals above the main diagonal
+ */
+ public AbstractBandMatrix(int n, int kl, int ku) {
+ super(n, n);
+
+ this.n = n;
+ if (kl < 0 || ku < 0)
+ throw new IllegalArgumentException("kl < 0 || ku < 0");
+ this.kl = kl;
+ this.ku = ku;
+
+ data = new double[numColumns * (1 + kl + ku)];
+ }
+
+ /**
+ * Constructor for AbstractBandMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param kl
+ * Number of diagonals below the main diagonal
+ * @param ku
+ * Number of diagonals above the main diagonal
+ */
+ public AbstractBandMatrix(Matrix A, int kl, int ku) {
+ this(A, kl, ku, true);
+ }
+
+ /**
+ * Constructor for AbstractBandMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param kl
+ * Number of diagonals below the main diagonal
+ * @param ku
+ * Number of diagonals above the main diagonal
+ * @param deep
+ * True if a deep copy is made. For a shallow copy,
+ * <code>A</code> must be a banded matrix
+ */
+ public AbstractBandMatrix(Matrix A, int kl, int ku, boolean deep) {
+ super(A);
+
+ if (kl < 0 || ku < 0)
+ throw new IllegalArgumentException("kl < 0 || ku < 0");
+ if (!isSquare())
+ throw new IllegalArgumentException("Band matrix must be square");
+ this.n = numRows;
+ this.kl = kl;
+ this.ku = ku;
+
+ if (deep) {
+ data = new double[numColumns * (1 + kl + ku)];
+ copy(A);
+ } else
+ this.data = ((AbstractBandMatrix) A).getData();
+ }
+
+ /**
+ * Returns the matrix contents
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ checkBand(row, column);
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ checkBand(row, column);
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (!inBand(row, column))
+ return 0;
+ return data[getIndex(row, column)];
+ }
+
+ /**
+ * Returns the number of lower diagonals
+ */
+ public int numSubDiagonals() {
+ return kl;
+ }
+
+ /**
+ * Returns the number of upper diagonals
+ */
+ public int numSuperDiagonals() {
+ return ku;
+ }
+
+ /**
+ * Returns true if the given indices are within the band
+ */
+ boolean inBand(int row, int column) {
+ return column - ku <= row && row <= column + kl;
+ }
+
+ /**
+ * Checks that the indices are within the band
+ */
+ void checkBand(int row, int column) {
+ if (!inBand(row, column))
+ throw new IndexOutOfBoundsException("Insertion index out of band");
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return ku + row - column + column * (kl + ku + 1);
+ }
+
+ /**
+ * Set this matrix equal to the given matrix
+ */
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (inBand(e.row(), e.column()))
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public Matrix set(Matrix B) {
+ if (!(B instanceof AbstractBandMatrix))
+ return super.set(B);
+
+ checkSize(B);
+
+ AbstractBandMatrix Bb = (AbstractBandMatrix) B;
+ if (Bb.kl != kl)
+ throw new IllegalArgumentException("B.kl != kl");
+ if (Bb.ku != ku)
+ throw new IllegalArgumentException("B.ku != ku");
+
+ double[] Bd = Bb.getData();
+
+ if (Bd == data)
+ return this;
+
+ System.arraycopy(Bd, 0, data, 0, data.length);
+
+ return this;
+ }
+
+ @Override
+ public Matrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new BandMatrixIterator();
+ }
+
+ /**
+ * Iterator over a band matrix
+ */
+ class BandMatrixIterator extends RefMatrixIterator {
+
+ /**
+ * Matrix bandwidths. Cannot be taken directly from the matrix since
+ * that breaks iterating over symmetrical matrices
+ */
+ private final int lkl, lku;
+
+ public BandMatrixIterator(int lkl, int lku) {
+ this.lkl = lkl;
+ this.lku = lku;
+ }
+
+ public BandMatrixIterator() {
+ this(kl, ku);
+ }
+
+ @Override
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ // Traversal first down the columns, then the rows
+ if (row < Math.min(column + lkl, n - 1)
+ && row >= Math.max(column - lku, 0))
+ row++;
+ else {
+ column++;
+ row = Math.max(column - lku, 0);
+ }
+
+ return entry;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractDenseMatrix.java
new file mode 100644
index 0000000..564ee0c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractDenseMatrix.java
@@ -0,0 +1,170 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.text.DecimalFormat;
+import java.util.Arrays;
+
+/**
+ * Partial implementation of a dense matrix
+ */
+abstract class AbstractDenseMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix contents
+ */
+ double[] data;
+
+ /**
+ * Constructor for AbstractDenseMatrix. The matrix contents will be set to
+ * zero
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of columns
+ */
+ public AbstractDenseMatrix(int numRows, int numColumns) {
+ super(numRows, numColumns);
+
+ // We know that numRows and numColumns is positive from the super
+ // constructor.
+
+ final long size = (long) numRows * numColumns;
+ if (size > Integer.MAX_VALUE)
+ {
+ throw new IllegalArgumentException("Matrix of "
+ + numRows + " x " + numColumns
+ + " = " + size + " elements is too large to be allocated using a single Java array.");
+ }
+
+ data = new double[numRows * numColumns];
+ }
+
+ /**
+ * Constructor for AbstractDenseMatrix. Matrix is copied from the supplied
+ * matrix
+ *
+ * @param A
+ * Matrix to copy from
+ */
+ public AbstractDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for AbstractDenseMatrix. Matrix is copied from the supplied
+ * matrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param deep
+ * True for deep copy, false for reference
+ */
+ public AbstractDenseMatrix(Matrix A, boolean deep) {
+ super(A);
+
+ if (deep) {
+ data = new double[numRows * numColumns];
+ copy(A);
+ } else
+ this.data = ((AbstractDenseMatrix) A).getData();
+ }
+
+ /**
+ * Set this matrix equal to the given matrix
+ */
+ abstract void copy(Matrix A);
+
+ /**
+ * Returns the matrix contents. Ordering depends on the underlying storage
+ * assumptions
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return data[getIndex(row, column)];
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + column * numRows;
+ }
+
+ @Override
+ public Matrix set(Matrix B) {
+ // using instanceof results in weird problems
+ // with implementations that mask some values
+ if (!(getClass().isAssignableFrom(B.getClass())))
+ return super.set(B);
+
+ checkSize(B);
+
+ double[] Bd = ((AbstractDenseMatrix) B).getData();
+
+ if (Bd == data)
+ return this;
+
+ System.arraycopy(Bd, 0, data, 0, data.length);
+
+ return this;
+ }
+
+ @Override
+ public Matrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ @Override
+ public String toString() {
+ StringBuilder out = new StringBuilder();
+ DecimalFormat df = new DecimalFormat("####0.00");
+
+ for (int i = 0; i < numRows(); i++) {
+ for (int j = 0; j < numColumns(); j++) {
+ double value = get(i, j);
+ if (value >= 0) out.append(" ");
+ out.append(" " + df.format(value));
+ }
+ out.append("\n");
+ }
+
+ return out.toString();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractMatrix.java
new file mode 100644
index 0000000..4e926a1
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractMatrix.java
@@ -0,0 +1,888 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Formatter;
+import java.util.Iterator;
+
+/**
+ * Partial implementation of <code>Matrix</code>. The following methods throw
+ * <code>UnsupportedOperationException</code>, and should be overridden by a
+ * subclass:
+ * <ul>
+ * <li><code>get(int,int)</code></li>
+ * <li><code>set(int,int,double)</code></li>
+ * <li><code>copy</code></li>
+ * <li>All the direct solution methods</li>
+ * </ul>
+ * <p>
+ * For the rest of the methods, simple default implementations using a matrix
+ * iterator has been provided. There are some kernel operations which the
+ * simpler operations forward to, for instance, <code>mult(Matrix,Matrix)</code>
+ * forwards to <code>multAdd(double,Matrix,Matrix)</code>. Subclasses can
+ * thus focus on overriding the kernel operations, which are:
+ * <ul>
+ * <li> <code>multAdd(double,Vector,Vector)</code> and
+ * <code>transMultAdd(double,Vector,Vector)</code>. </li>
+ * <li> <code>rank1(double,Vector,Vector)</code> and
+ * <code>rank1(double,Vector,Vector)</code>.</li>
+ * <li> <code>multAdd(double,Matrix,Matrix)</code>,
+ * <code>transAmultAdd(double,Matrix,Matrix)</code>,
+ * <code>transBmultAdd(double,Matrix,Matrix)</code>, and
+ * <code>transABmultAdd(double,Matrix,Matrix)</code>. </li>
+ * <li> <code>scale(double)</code>. </li>
+ * <li> <code>set(double,Matrix)</code> and <code>add(double,Matrix)</code>.
+ * </li>
+ * <li> <code>transpose</code> and <code>transpose(Matrix)</code>. </li>
+ * <li> All the norms.</li>
+ * </ul>
+ * <p>
+ * Finally, a default iterator is provided by this class, which works by calling
+ * the <code>get</code> function. A tailored replacement should be used by
+ * subclasses.
+ * </ul>
+ */
+public abstract class AbstractMatrix implements Matrix {
+
+ /**
+ * Number of rows
+ */
+ protected int numRows;
+
+ /**
+ * Number of columns
+ */
+ protected int numColumns;
+
+ /**
+ * Constructor for AbstractMatrix
+ */
+ protected AbstractMatrix(int numRows, int numColumns) {
+ if (numRows < 0 || numColumns < 0)
+ throw new IndexOutOfBoundsException(
+ "Matrix size cannot be negative");
+ this.numRows = numRows;
+ this.numColumns = numColumns;
+ }
+
+ /**
+ * Constructor for AbstractMatrix, same size as A. The invoking constructor
+ * should set this matrix equal the argument matrix
+ */
+ protected AbstractMatrix(Matrix A) {
+ this(A.numRows(), A.numColumns());
+ }
+
+ public int numRows() {
+ return numRows;
+ }
+
+ public int numColumns() {
+ return numColumns;
+ }
+
+ public boolean isSquare() {
+ return numRows == numColumns;
+ }
+
+ public void set(int row, int column, double value) {
+ throw new UnsupportedOperationException();
+ }
+
+ public void add(int row, int column, double value) {
+ set(row, column, value + get(row, column));
+ }
+
+ public double get(int row, int column) {
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Checks the passed row and column indices
+ */
+ protected void check(int row, int column) {
+ if (row < 0)
+ throw new IndexOutOfBoundsException("row index is negative (" + row
+ + ")");
+ if (column < 0)
+ throw new IndexOutOfBoundsException("column index is negative ("
+ + column + ")");
+ if (row >= numRows)
+ throw new IndexOutOfBoundsException("row index >= numRows (" + row
+ + " >= " + numRows + ")");
+ if (column >= numColumns)
+ throw new IndexOutOfBoundsException("column index >= numColumns ("
+ + column + " >= " + numColumns + ")");
+ }
+
+ public Matrix copy() {
+ throw new UnsupportedOperationException();
+ }
+
+ public Matrix zero() {
+ for (MatrixEntry e : this)
+ e.set(0);
+ return this;
+ }
+
+ public Vector mult(Vector x, Vector y) {
+ return mult(1, x, y);
+ }
+
+ public Vector mult(double alpha, Vector x, Vector y) {
+ return multAdd(alpha, x, y.zero());
+ }
+
+ public Vector multAdd(Vector x, Vector y) {
+ return multAdd(1, x, y);
+ }
+
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ checkMultAdd(x, y);
+
+ if (alpha != 0)
+ for (MatrixEntry e : this)
+ y.add(e.row(), alpha * e.get() * x.get(e.column()));
+
+ return y;
+ }
+
+ /**
+ * Checks the arguments to <code>mult</code> and <code>multAdd</code>
+ */
+ protected void checkMultAdd(Vector x, Vector y) {
+ if (numColumns != x.size())
+ throw new IndexOutOfBoundsException("A.numColumns != x.size ("
+ + numColumns + " != " + x.size() + ")");
+ if (numRows != y.size())
+ throw new IndexOutOfBoundsException("A.numRows != y.size ("
+ + numRows + " != " + y.size() + ")");
+ }
+
+ public Vector transMult(Vector x, Vector y) {
+ return transMult(1, x, y);
+ }
+
+ public Vector transMult(double alpha, Vector x, Vector y) {
+ return transMultAdd(alpha, x, y.zero());
+ }
+
+ public Vector transMultAdd(Vector x, Vector y) {
+ return transMultAdd(1, x, y);
+ }
+
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ checkTransMultAdd(x, y);
+
+ if (alpha != 0)
+ for (MatrixEntry e : this)
+ y.add(e.column(), alpha * e.get() * x.get(e.row()));
+
+ return y;
+ }
+
+ /**
+ * Checks the arguments to <code>transMult</code> and
+ * <code>transMultAdd</code>
+ */
+ protected void checkTransMultAdd(Vector x, Vector y) {
+ if (numRows != x.size())
+ throw new IndexOutOfBoundsException("A.numRows != x.size ("
+ + numRows + " != " + x.size() + ")");
+ if (numColumns != y.size())
+ throw new IndexOutOfBoundsException("A.numColumns != y.size ("
+ + numColumns + " != " + y.size() + ")");
+ }
+
+ public Vector solve(Vector b, Vector x) {
+ throw new UnsupportedOperationException();
+ }
+
+ public Vector transSolve(Vector b, Vector x) {
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Checks that a matrix inversion is legal for the given arguments. This is
+ * for the square case, not for least-squares problems
+ */
+ protected void checkSolve(Vector b, Vector x) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (numRows != b.size())
+ throw new IndexOutOfBoundsException("numRows != b.size (" + numRows
+ + " != " + b.size() + ")");
+ if (numColumns != x.size())
+ throw new IndexOutOfBoundsException("numColumns != x.size ("
+ + numColumns + " != " + x.size() + ")");
+ }
+
+ public Matrix rank1(Vector x) {
+ return rank1(1, x);
+ }
+
+ public Matrix rank1(double alpha, Vector x) {
+ return rank1(alpha, x, x);
+ }
+
+ public Matrix rank1(Vector x, Vector y) {
+ return rank1(1, x, y);
+ }
+
+ public Matrix rank1(double alpha, Vector x, Vector y) {
+ checkRank1(x, y);
+
+ if (alpha == 0)
+ return this;
+
+ for (VectorEntry ei : x)
+ if (ei.get() != 0)
+ for (VectorEntry ej : y)
+ if (ej.get() != 0)
+ add(ei.index(), ej.index(), alpha * ei.get() * ej.get());
+
+ return this;
+ }
+
+ /**
+ * Checks that a vector rank1 update is possible for the given vectors
+ */
+ protected void checkRank1(Vector x, Vector y) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (x.size() != numRows)
+ throw new IndexOutOfBoundsException("x.size != A.numRows ("
+ + x.size() + " != " + numRows + ")");
+ if (y.size() != numColumns)
+ throw new IndexOutOfBoundsException("y.size != A.numColumns ("
+ + y.size() + " != " + numColumns + ")");
+ }
+
+ public Matrix rank2(Vector x, Vector y) {
+ return rank2(1, x, y);
+ }
+
+ public Matrix rank2(double alpha, Vector x, Vector y) {
+ checkRank2(x, y);
+
+ if (alpha == 0)
+ return this;
+
+ for (VectorEntry ei : x)
+ for (VectorEntry ej : y) {
+ add(ei.index(), ej.index(), alpha * ei.get() * ej.get());
+ add(ej.index(), ei.index(), alpha * ei.get() * ej.get());
+ }
+
+ return this;
+ }
+
+ /**
+ * Checks that a vector rank2 update is legal with the given vectors
+ */
+ protected void checkRank2(Vector x, Vector y) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (x.size() != numRows)
+ throw new IndexOutOfBoundsException("x.size != A.numRows ("
+ + x.size() + " != " + numRows + ")");
+ if (y.size() != numRows)
+ throw new IndexOutOfBoundsException("y.size != A.numRows ("
+ + y.size() + " != " + numRows + ")");
+ }
+
+ public Matrix mult(Matrix B, Matrix C) {
+ return mult(1, B, C);
+ }
+
+ public Matrix mult(double alpha, Matrix B, Matrix C) {
+ return multAdd(alpha, B, C.zero());
+ }
+
+ public Matrix multAdd(Matrix B, Matrix C) {
+ return multAdd(1, B, C);
+ }
+
+ public Matrix multAdd(double alpha, Matrix B, Matrix C) {
+ checkMultAdd(B, C);
+
+ if (alpha != 0)
+ for (int i = 0; i < numRows; ++i)
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = 0; k < numColumns; ++k)
+ dot += get(i, k) * B.get(k, j);
+ C.add(i, j, alpha * dot);
+ }
+
+ return C;
+ }
+
+ /**
+ * Checks the arguments to <code>mult</code> and <code>multAdd</code>
+ */
+ protected void checkMultAdd(Matrix B, Matrix C) {
+ if (numRows != C.numRows())
+ throw new IndexOutOfBoundsException("A.numRows != C.numRows ("
+ + numRows + " != " + C.numRows() + ")");
+ if (numColumns != B.numRows())
+ throw new IndexOutOfBoundsException("A.numColumns != B.numRows ("
+ + numColumns + " != " + B.numRows() + ")");
+ if (B.numColumns() != C.numColumns())
+ throw new IndexOutOfBoundsException(
+ "B.numColumns != C.numColumns (" + B.numRows() + " != "
+ + C.numColumns() + ")");
+ }
+
+ public Matrix transAmult(Matrix B, Matrix C) {
+ return transAmult(1, B, C);
+ }
+
+ public Matrix transAmult(double alpha, Matrix B, Matrix C) {
+ return transAmultAdd(alpha, B, C.zero());
+ }
+
+ public Matrix transAmultAdd(Matrix B, Matrix C) {
+ return transAmultAdd(1, B, C);
+ }
+
+ public Matrix transAmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransAmultAdd(B, C);
+
+ if (alpha != 0)
+ for (int i = 0; i < numColumns; ++i)
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = 0; k < numRows; ++k)
+ dot += get(k, i) * B.get(k, j);
+ C.add(i, j, alpha * dot);
+ }
+
+ return C;
+ }
+
+ /**
+ * Checks the arguments to <code>transAmult</code> and
+ * <code>transAmultAdd</code>
+ */
+ protected void checkTransAmultAdd(Matrix B, Matrix C) {
+ if (numRows != B.numRows())
+ throw new IndexOutOfBoundsException("A.numRows != B.numRows ("
+ + numRows + " != " + B.numRows() + ")");
+ if (numColumns != C.numRows())
+ throw new IndexOutOfBoundsException("A.numColumns != C.numRows ("
+ + numColumns + " != " + C.numRows() + ")");
+ if (B.numColumns() != C.numColumns())
+ throw new IndexOutOfBoundsException(
+ "B.numColumns != C.numColumns (" + B.numColumns() + " != "
+ + C.numColumns() + ")");
+ }
+
+ public Matrix transBmult(Matrix B, Matrix C) {
+ return transBmult(1, B, C);
+ }
+
+ public Matrix transBmult(double alpha, Matrix B, Matrix C) {
+ return transBmultAdd(alpha, B, C.zero());
+ }
+
+ public Matrix transBmultAdd(Matrix B, Matrix C) {
+ return transBmultAdd(1, B, C);
+ }
+
+ public Matrix transBmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransBmultAdd(B, C);
+
+ if (alpha != 0)
+ for (int i = 0; i < numRows; ++i)
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = 0; k < numColumns; ++k)
+ dot += get(i, k) * B.get(j, k);
+ C.add(i, j, alpha * dot);
+ }
+
+ return C;
+ }
+
+ /**
+ * Checks the arguments to <code>transBmult</code> and
+ * <code>transBmultAdd</code>
+ */
+ protected void checkTransBmultAdd(Matrix B, Matrix C) {
+ if (numColumns != B.numColumns())
+ throw new IndexOutOfBoundsException(
+ "A.numColumns != B.numColumns (" + numColumns + " != "
+ + B.numColumns() + ")");
+ if (numRows != C.numRows())
+ throw new IndexOutOfBoundsException("A.numRows != C.numRows ("
+ + numRows + " != " + C.numRows() + ")");
+ if (B.numRows() != C.numColumns())
+ throw new IndexOutOfBoundsException("B.numRows != C.numColumns ("
+ + B.numRows() + " != " + C.numColumns() + ")");
+ }
+
+ public Matrix transABmult(Matrix B, Matrix C) {
+ return transABmult(1, B, C);
+ }
+
+ public Matrix transABmult(double alpha, Matrix B, Matrix C) {
+ return transABmultAdd(alpha, B, C.zero());
+ }
+
+ public Matrix transABmultAdd(Matrix B, Matrix C) {
+ return transABmultAdd(1, B, C);
+ }
+
+ public Matrix transABmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransABmultAdd(B, C);
+
+ if (alpha != 0)
+ for (int i = 0; i < numColumns; ++i)
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = 0; k < numRows; ++k)
+ dot += get(k, i) * B.get(j, k);
+ C.add(i, j, alpha * dot);
+ }
+
+ return C;
+ }
+
+ /**
+ * Checks the arguments to <code>transABmultAdd</code> and
+ * <code>transABmultAdd</code>
+ */
+ protected void checkTransABmultAdd(Matrix B, Matrix C) {
+ if (numRows != B.numColumns())
+ throw new IndexOutOfBoundsException("A.numRows != B.numColumns ("
+ + numRows + " != " + B.numColumns() + ")");
+ if (numColumns != C.numRows())
+ throw new IndexOutOfBoundsException("A.numColumns != C.numRows ("
+ + numColumns + " != " + C.numRows() + ")");
+ if (B.numRows() != C.numColumns())
+ throw new IndexOutOfBoundsException("B.numRows != C.numColumns ("
+ + B.numRows() + " != " + C.numColumns() + ")");
+ }
+
+ public Matrix solve(Matrix B, Matrix X) {
+ throw new UnsupportedOperationException();
+ }
+
+ public Matrix transSolve(Matrix B, Matrix X) {
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Checks that a matrix inversion is legal for the given arguments. This is
+ * for the square case, not for least-squares problems
+ */
+ protected void checkSolve(Matrix B, Matrix X) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (B.numRows() != numRows)
+ throw new IndexOutOfBoundsException("B.numRows != A.numRows ("
+ + B.numRows() + " != " + numRows + ")");
+ if (B.numColumns() != X.numColumns())
+ throw new IndexOutOfBoundsException(
+ "B.numColumns != X.numColumns (" + B.numColumns() + " != "
+ + X.numColumns() + ")");
+ if (X.numRows() != numColumns)
+ throw new IndexOutOfBoundsException("X.numRows != A.numColumns ("
+ + X.numRows() + " != " + numColumns + ")");
+ }
+
+ public Matrix rank1(Matrix C) {
+ return rank1(1, C);
+ }
+
+ public Matrix rank1(double alpha, Matrix C) {
+ checkRank1(C);
+
+ if (alpha == 0)
+ return this;
+
+ return C.transBmultAdd(alpha, C, this);
+ }
+
+ /**
+ * Checks that a matrix rank1 update is possible for the given matrix
+ */
+ protected void checkRank1(Matrix C) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (numRows != C.numRows())
+ throw new IndexOutOfBoundsException("A.numRows != C.numRows ("
+ + numRows + " != " + C.numRows() + ")");
+ }
+
+ public Matrix transRank1(Matrix C) {
+ return transRank1(1, C);
+ }
+
+ public Matrix transRank1(double alpha, Matrix C) {
+ checkTransRank1(C);
+
+ if (alpha == 0)
+ return this;
+
+ return C.transAmultAdd(alpha, C, this);
+ }
+
+ /**
+ * Checks that a transposed rank1 update is leagal with the given argument
+ */
+ protected void checkTransRank1(Matrix C) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (numRows != C.numColumns())
+ throw new IndexOutOfBoundsException("A.numRows != C.numColumns ("
+ + numRows + " != " + C.numColumns() + ")");
+ }
+
+ public Matrix rank2(Matrix B, Matrix C) {
+ return rank2(1, B, C);
+ }
+
+ public Matrix rank2(double alpha, Matrix B, Matrix C) {
+ checkRank2(B, C);
+
+ if (alpha == 0)
+ return this;
+
+ return B.transBmultAdd(alpha, C, C.transBmultAdd(alpha, B, this));
+ }
+
+ /**
+ * Checks that a rank2 update is legal for the given arguments
+ */
+ protected void checkRank2(Matrix B, Matrix C) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (B.numRows() != C.numRows())
+ throw new IndexOutOfBoundsException("B.numRows != C.numRows ("
+ + B.numRows() + " != " + C.numRows() + ")");
+ if (B.numColumns() != C.numColumns())
+ throw new IndexOutOfBoundsException(
+ "B.numColumns != C.numColumns (" + B.numColumns() + " != "
+ + C.numColumns() + ")");
+ }
+
+ public Matrix transRank2(Matrix B, Matrix C) {
+ return transRank2(1, B, C);
+ }
+
+ public Matrix transRank2(double alpha, Matrix B, Matrix C) {
+ checkTransRank2(B, C);
+
+ if (alpha == 0)
+ return this;
+
+ return B.transAmultAdd(alpha, C, C.transAmultAdd(alpha, B, this));
+ }
+
+ /**
+ * Checks that a transposed rank2 update is leagal with the given arguments
+ */
+ protected void checkTransRank2(Matrix B, Matrix C) {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ if (numRows != B.numColumns())
+ throw new IndexOutOfBoundsException("A.numRows != B.numColumns ("
+ + numRows + " != " + B.numColumns() + ")");
+ if (B.numRows() != C.numRows())
+ throw new IndexOutOfBoundsException("B.numRows != C.numRows ("
+ + B.numRows() + " != " + C.numRows() + ")");
+ if (B.numColumns() != C.numColumns())
+ throw new IndexOutOfBoundsException(
+ "B.numColumns != C.numColumns (" + B.numColumns() + " != "
+ + C.numColumns() + ")");
+ }
+
+ public Matrix scale(double alpha) {
+ if (alpha == 1)
+ return this;
+ else if (alpha == 0)
+ return zero();
+
+ for (MatrixEntry e : this)
+ e.set(alpha * e.get());
+
+ return this;
+ }
+
+ public Matrix set(Matrix B) {
+ return set(1, B);
+ }
+
+ public Matrix set(double alpha, Matrix B) {
+ checkSize(B);
+
+ if (alpha == 0.)
+ return zero();
+ if (B == this)
+ return scale(alpha);
+
+ zero();
+ for (MatrixEntry e : B)
+ if (e.get() != 0) set(e.row(), e.column(), alpha * e.get());
+
+ return this;
+ }
+
+ public Matrix add(Matrix B) {
+ return add(1, B);
+ }
+
+ public Matrix add(double alpha, Matrix B) {
+ checkSize(B);
+
+ if (alpha != 0)
+ for (MatrixEntry e : B)
+ add(e.row(), e.column(), alpha * e.get());
+
+ return this;
+ }
+
+ /**
+ * Checks that the sizes of this matrix and the given conform
+ */
+ protected void checkSize(Matrix B) {
+ if (numRows != B.numRows())
+ throw new IndexOutOfBoundsException("A.numRows != B.numRows ("
+ + numRows + " != " + B.numRows() + ")");
+ if (numColumns != B.numColumns())
+ throw new IndexOutOfBoundsException(
+ "A.numColumns != B.numColumns (" + numColumns + " != "
+ + B.numColumns() + ")");
+ }
+
+ public Matrix transpose() {
+ checkTranspose();
+
+ for (int j = 0; j < numColumns; ++j)
+ for (int i = j + 1; i < numRows; ++i) {
+ double value = get(i, j);
+ set(i, j, get(j, i));
+ set(j, i, value);
+ }
+
+ return this;
+ }
+
+ /**
+ * Checks that the matrix may be transposed
+ */
+ protected void checkTranspose() {
+ if (!isSquare())
+ throw new IndexOutOfBoundsException("!A.isSquare");
+ }
+
+ public Matrix transpose(Matrix B) {
+ checkTranspose(B);
+
+ if (B == this)
+ return transpose();
+
+ B.zero();
+ for (MatrixEntry e : this)
+ B.set(e.column(), e.row(), e.get());
+
+ return B;
+ }
+
+ /**
+ * Checks that this matrix can be transposed into the given matrix
+ */
+ protected void checkTranspose(Matrix B) {
+ if (numRows != B.numColumns())
+ throw new IndexOutOfBoundsException("A.numRows != B.numColumns ("
+ + numRows + " != " + B.numColumns() + ")");
+ if (numColumns != B.numRows())
+ throw new IndexOutOfBoundsException("A.numColumns != B.numRows ("
+ + numColumns + " != " + B.numRows() + ")");
+ }
+
+ public double norm(Norm type) {
+ if (type == Norm.One)
+ return norm1();
+ else if (type == Norm.Frobenius)
+ return normF();
+ else if (type == Norm.Infinity)
+ return normInf();
+ else
+ // Maxvalue
+ return max();
+ }
+
+ /**
+ * Computes the 1 norm
+ */
+ protected double norm1() {
+ double[] rowSum = new double[numRows];
+ for (MatrixEntry e : this)
+ rowSum[e.row()] += Math.abs(e.get());
+ return max(rowSum);
+ }
+
+ /**
+ * Computes the Frobenius norm. This implementation is overflow resistant
+ */
+ protected double normF() {
+ double scale = 0, ssq = 1;
+ for (MatrixEntry e : this) {
+ double Aval = e.get();
+ if (Aval != 0) {
+ double absxi = Math.abs(Aval);
+ if (scale < absxi) {
+ ssq = 1 + ssq * Math.pow(scale / absxi, 2);
+ scale = absxi;
+ } else
+ ssq = ssq + Math.pow(absxi / scale, 2);
+ }
+ }
+ return scale * Math.sqrt(ssq);
+ }
+
+ /**
+ * Computes the infinity norm
+ */
+ protected double normInf() {
+ double[] columnSum = new double[numColumns];
+ for (MatrixEntry e : this)
+ columnSum[e.column()] += Math.abs(e.get());
+ return max(columnSum);
+ }
+
+ /**
+ * Returns the largest absolute value
+ */
+ protected double max() {
+ double max = 0;
+ for (MatrixEntry e : this)
+ max = Math.max(Math.abs(e.get()), max);
+ return max;
+ }
+
+ /**
+ * Returns the largest element of the passed array
+ */
+ protected double max(double[] x) {
+ double max = 0;
+ for (int i = 0; i < x.length; ++i)
+ max = Math.max(x[i], max);
+ return max;
+ }
+
+ @Override
+ public String toString() {
+ // Output into coordinate format. Indices start from 1 instead of 0
+ Formatter out = new Formatter();
+
+ out.format("%10d %10d %19d\n", numRows, numColumns, Matrices
+ .cardinality(this));
+
+ for (MatrixEntry e : this)
+ if (e.get() != 0)
+ out.format("%10d %10d % .12e\n", e.row() + 1, e.column() + 1, e.get());
+
+ return out.toString();
+ }
+
+ public Iterator<MatrixEntry> iterator() {
+ return new RefMatrixIterator();
+ }
+
+ /**
+ * Iterator over a general matrix. Uses column-major traversal
+ */
+ class RefMatrixIterator implements Iterator<MatrixEntry> {
+
+ /**
+ * Matrix cursor
+ */
+ int row, column;
+
+ /**
+ * Matrix entry
+ */
+ final RefMatrixEntry entry = new RefMatrixEntry();
+
+ public boolean hasNext() {
+ return (row < numRows) && (column < numColumns);
+ }
+
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ // Traversal first down the columns, then the rows
+ if (row < numRows - 1)
+ row++;
+ else {
+ column++;
+ row = 0;
+ }
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Matrix entry backed by the matrix. May be reused for higher performance
+ */
+ class RefMatrixEntry implements MatrixEntry {
+
+ /**
+ * Matrix position
+ */
+ private int row, column;
+
+ /**
+ * Updates the entry
+ */
+ public void update(int row, int column) {
+ this.row = row;
+ this.column = column;
+ }
+
+ public int row() {
+ return row;
+ }
+
+ public int column() {
+ return column;
+ }
+
+ public double get() {
+ return AbstractMatrix.this.get(row, column);
+ }
+
+ public void set(double value) {
+ AbstractMatrix.this.set(row, column, value);
+ }
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractPackMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractPackMatrix.java
new file mode 100644
index 0000000..5d5ed7f
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractPackMatrix.java
@@ -0,0 +1,121 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+
+/**
+ * Partial implementation of a packed matrix
+ */
+abstract class AbstractPackMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix contents
+ */
+ double[] data;
+
+ /**
+ * Matrix is square, so this is either numRows or numColumns
+ */
+ int n;
+
+ /**
+ * Constructor for AbstractPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public AbstractPackMatrix(int n) {
+ super(n, n);
+ this.n = numRows;
+ data = new double[(n * n + n) / 2];
+ }
+
+ /**
+ * Constructor for AbstractPackMatrix
+ *
+ * @param A
+ * Matrix to copy
+ */
+ public AbstractPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for AbstractPackMatrix
+ *
+ * @param A
+ * A square matrix to copy from
+ * @param deep
+ * True for a deep copy, false for a shallow (reference) copy.
+ * References must be instances of a packed matrix
+ */
+ public AbstractPackMatrix(Matrix A, boolean deep) {
+ super(A);
+
+ if (!isSquare())
+ throw new IllegalArgumentException("Packed matrix must be square");
+ n = A.numRows();
+ if (deep) {
+ data = new double[(n * n + n) / 2];
+ copy(A);
+ } else
+ this.data = ((AbstractPackMatrix) A).getData();
+ }
+
+ /**
+ * Set this matrix equal to the given matrix
+ */
+ abstract void copy(Matrix A);
+
+ /**
+ * Returns the matrix contents. Ordering depends on the underlying storage
+ * assumptions
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ @Override
+ public Matrix set(Matrix B) {
+ if (!(B instanceof AbstractPackMatrix))
+ return super.set(B);
+
+ checkSize(B);
+
+ double[] Bd = ((AbstractPackMatrix) B).getData();
+
+ if (Bd == data)
+ return this;
+
+ System.arraycopy(Bd, 0, data, 0, data.length);
+
+ return this;
+ }
+
+ @Override
+ public Matrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractSymmBandMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractSymmBandMatrix.java
new file mode 100644
index 0000000..aeccfdb
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractSymmBandMatrix.java
@@ -0,0 +1,168 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Iterator;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Partial implementation of a symmetrical, banded matrix
+ */
+abstract class AbstractSymmBandMatrix extends AbstractBandMatrix {
+
+ /**
+ * Upper or lower part stored?
+ */
+ private UpLo uplo;
+
+ /**
+ * Diagonals in relevant band
+ */
+ int kd;
+
+ /**
+ * Constructor for AbstractSymmBandMatrix
+ */
+ AbstractSymmBandMatrix(int n, int kl, int ku, UpLo uplo) {
+ super(n, kl, ku);
+ kd = Math.max(kl, ku);
+ this.uplo = uplo;
+ }
+
+ /**
+ * Constructor for AbstractSymmBandMatrix
+ */
+ AbstractSymmBandMatrix(Matrix A, int kl, int ku, UpLo uplo) {
+ this(A, kl, ku, true, uplo);
+ }
+
+ /**
+ * Constructor for AbstractSymmBandMatrix
+ */
+ AbstractSymmBandMatrix(Matrix A, int kl, int ku, boolean deep, UpLo uplo) {
+ super(A, kl, ku, deep);
+ kd = Math.max(kl, ku);
+ this.uplo = uplo;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dsbmv(uplo.netlib(), numRows, kd, alpha, data, kd + 1, xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ return multAdd(alpha, x, y);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new BandMatrixIterator(kd, kd);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ // Allocate factorization matrix. The factorization matrix will be
+ // large enough to accomodate any pivots
+ BandMatrix Af = new BandMatrix(this, kd, kd + kd);
+ int[] ipiv = new int[numRows];
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgbsv(numRows, kd, kd, X.numColumns(),
+ Af.getData(), Matrices.ld(2 * kd + kd + 1), ipiv, Xd,
+ Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ return solve(b, x);
+ }
+
+ Matrix SPDsolve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dpbsv(uplo.netlib(), numRows, kd, X.numColumns(),
+ data.clone(), Matrices.ld(kd + 1), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixNotSPDException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Matrix transpose() {
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractSymmDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractSymmDenseMatrix.java
new file mode 100644
index 0000000..6df0e15
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractSymmDenseMatrix.java
@@ -0,0 +1,291 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Partial implementation of a symmetrical, dense matrix
+ */
+abstract class AbstractSymmDenseMatrix extends AbstractDenseMatrix {
+
+ /**
+ * Upper or lower part stored?
+ */
+ private UpLo uplo;
+
+ /**
+ * Constructor for AbstractSymmDenseMatrix
+ */
+ AbstractSymmDenseMatrix(int n, UpLo uplo) {
+ super(n, n);
+ this.uplo = uplo;
+ }
+
+ /**
+ * Constructor for AbstractSymmDenseMatrix
+ */
+ AbstractSymmDenseMatrix(Matrix A, UpLo uplo) {
+ this(A, true, uplo);
+ }
+
+ /**
+ * Constructor for AbstractSymmDenseMatrix
+ */
+ AbstractSymmDenseMatrix(Matrix A, boolean deep, UpLo uplo) {
+ super(A, deep);
+ if (!isSquare())
+ throw new IllegalArgumentException(
+ "Symmetric matrix must be square");
+ this.uplo = uplo;
+ }
+
+ @Override
+ public Matrix multAdd(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.multAdd(alpha, B, C);
+
+ checkMultAdd(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dsymm(Side.Left.netlib(), uplo.netlib(), C.numRows(), C.numColumns(),
+ alpha, data, Math.max(1, C.numRows()), Bd,
+ Math.max(1, C.numRows()), 1, Cd, Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix transAmultAdd(double alpha, Matrix B, Matrix C) {
+ return multAdd(alpha, B, C);
+ }
+
+ @Override
+ public Matrix rank1(double alpha, Vector x, Vector y) {
+ if (x != y)
+ throw new IllegalArgumentException("x != y");
+ if (!(x instanceof DenseVector))
+ return super.rank1(alpha, x, y);
+
+ checkRank1(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ BLAS.getInstance().dsyr(uplo.netlib(), numRows, alpha, xd, 1, data,
+ Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Matrix rank2(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.rank2(alpha, x, y);
+
+ checkRank2(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dsyr2(uplo.netlib(), numRows, alpha, xd, 1, yd, 1, data,
+ Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dsymv(uplo.netlib(), numRows, alpha, data, Math.max(1, numRows),
+ xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ return multAdd(alpha, x, y);
+ }
+
+ @Override
+ public Matrix rank1(double alpha, Matrix C) {
+ if (!(C instanceof DenseMatrix))
+ return super.rank1(alpha, C);
+
+ checkRank1(C);
+
+ double[] Cd = ((DenseMatrix) C).getData();
+
+ BLAS.getInstance().dsyrk(uplo.netlib(), Transpose.NoTranspose.netlib(), numRows,
+ C.numColumns(), alpha, Cd, Math.max(1, numRows), 1, data,
+ Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Matrix transRank1(double alpha, Matrix C) {
+ if (!(C instanceof DenseMatrix))
+ return super.transRank1(alpha, C);
+
+ checkTransRank1(C);
+
+ double[] Cd = ((DenseMatrix) C).getData();
+
+ BLAS.getInstance().dsyrk(uplo.netlib(), Transpose.Transpose.netlib(), numRows, numRows,
+ alpha, Cd, Math.max(1, numRows), 1, data, Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Matrix rank2(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.rank2(alpha, B, C);
+
+ checkRank2(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dsyr2k(uplo.netlib(), Transpose.NoTranspose.netlib(), numRows,
+ B.numColumns(), alpha, Bd, Math.max(1, numRows), Cd,
+ Math.max(1, numRows), 1, data, Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Matrix transRank2(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.transRank2(alpha, B, C);
+
+ checkTransRank2(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dsyr2k(uplo.netlib(), Transpose.Transpose.netlib(), numRows, B.numRows(),
+ alpha, Bd, Math.max(1, B.numRows()), Cd,
+ Math.max(1, B.numRows()), 1, data, Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ // Allocate factorization matrix
+ double[] newData = data.clone();
+ int[] ipiv = new int[numRows];
+
+ // Query optimal workspace
+ double[] work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dsysv(uplo.netlib(), numRows, X.numColumns(),
+ newData, Matrices.ld(numRows), ipiv, Xd, Matrices.ld(numRows),
+ work, -1, info);
+
+ // Allocate workspace
+ int lwork = -1;
+ if (info.val != 0)
+ lwork = 1;
+ else
+ lwork = Math.max((int) work[0], 1);
+ work = new double[lwork];
+
+ // Solve
+ info.val = 0;
+ LAPACK.getInstance().dsysv(uplo.netlib(), numRows, X.numColumns(), newData,
+ Matrices.ld(numRows), ipiv, Xd, Matrices.ld(numRows), work, lwork, info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ return solve(b, x);
+ }
+
+ Matrix SPDsolve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dposv(uplo.netlib(), numRows, X.numColumns(),
+ data.clone(), Matrices.ld(numRows), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixNotSPDException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Matrix transpose() {
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractSymmPackMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractSymmPackMatrix.java
new file mode 100644
index 0000000..6548f70
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractSymmPackMatrix.java
@@ -0,0 +1,180 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Partial implementation of a symmetrical, packed matrix
+ */
+abstract class AbstractSymmPackMatrix extends AbstractPackMatrix {
+
+ /**
+ * Which part of the matrix which is stored
+ */
+ private UpLo uplo;
+
+ /**
+ * Constructor for AbstractSymmPackMatrix
+ */
+ AbstractSymmPackMatrix(int n, UpLo uplo) {
+ super(n);
+ this.uplo = uplo;
+ }
+
+ /**
+ * Constructor for AbstractSymmPackMatrix
+ */
+ AbstractSymmPackMatrix(Matrix A, UpLo uplo) {
+ this(A, true, uplo);
+ }
+
+ /**
+ * Constructor for AbstractSymmPackMatrix
+ */
+ AbstractSymmPackMatrix(Matrix A, boolean deep, UpLo uplo) {
+ super(A, deep);
+ this.uplo = uplo;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dspmv(uplo.netlib(), numRows, alpha, data, xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ return multAdd(alpha, x, y);
+ }
+
+ @Override
+ public Matrix rank1(double alpha, Vector x, Vector y) {
+ if (x != y)
+ throw new IllegalArgumentException("x != y");
+ if (!(x instanceof DenseVector))
+ return super.rank1(alpha, x, y);
+
+ checkRank1(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ BLAS.getInstance().dspr(uplo.netlib(), numRows, alpha, xd, 1, data);
+
+ return this;
+ }
+
+ @Override
+ public Matrix rank2(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.rank2(alpha, x, y);
+
+ checkRank2(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dspr2(uplo.netlib(), numRows, alpha, xd, 1, yd, 1, data);
+
+ return this;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ int[] ipiv = new int[numRows];
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dspsv(uplo.netlib(), numRows, X.numColumns(),
+ data.clone(), ipiv, Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ return solve(b, x);
+ }
+
+ Matrix SPDsolve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dppsv(uplo.netlib(), numRows, X.numColumns(),
+ data.clone(), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixNotSPDException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Matrix transpose() {
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractTriangBandMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractTriangBandMatrix.java
new file mode 100644
index 0000000..6dafa48
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractTriangBandMatrix.java
@@ -0,0 +1,160 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Partial implementation of a triangular, banded matrix
+ */
+abstract class AbstractTriangBandMatrix extends AbstractBandMatrix {
+
+ /**
+ * Upper or lower part stored?
+ */
+ private UpLo uplo;
+
+ /**
+ * Diagonal stored or not?
+ */
+ private Diag diag;
+
+ /**
+ * Diagonals in relevant band
+ */
+ int kd;
+
+ /**
+ * Constructor for AbstractTriangBandMatrix
+ */
+ AbstractTriangBandMatrix(int n, int kl, int ku, UpLo uplo, Diag diag) {
+ super(n, kl, ku);
+ kd = Math.max(kl, ku);
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ /**
+ * Constructor for AbstractTriangBandMatrix
+ */
+ AbstractTriangBandMatrix(Matrix A, int kl, int ku, UpLo uplo, Diag diag) {
+ this(A, kl, ku, true, uplo, diag);
+ }
+
+ /**
+ * Constructor for AbstractTriangBandMatrix
+ */
+ AbstractTriangBandMatrix(Matrix A, int kl, int ku, boolean deep, UpLo uplo,
+ Diag diag) {
+ super(A, kl, ku, deep);
+ kd = Math.max(kl, ku);
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ @Override
+ public Vector mult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.mult(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A*y
+ BLAS.getInstance().dtbmv(uplo.netlib(), Transpose.NoTranspose.netlib(), diag.netlib(),
+ numRows, kd, data, kd + 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.transMult(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A*y
+ BLAS.getInstance().dtbmv(uplo.netlib(), Transpose.Transpose.netlib(), diag.netlib(),
+ numRows, kd, data, kd + 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.NoTranspose);
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.Transpose);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ transSolve(B, X);
+ return x;
+ }
+
+ Matrix solve(Matrix B, Matrix X, Transpose trans) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dtbtrs(uplo.netlib(), trans.netlib(), diag.netlib(),
+ numRows, kd, X.numColumns(), data, Matrices.ld(kd + 1), Xd, Matrices.ld(n),
+ info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractTriangDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractTriangDenseMatrix.java
new file mode 100644
index 0000000..364f144
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractTriangDenseMatrix.java
@@ -0,0 +1,295 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Iterator;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Partial implementation of a triangular, dense matrix
+ */
+abstract class AbstractTriangDenseMatrix extends AbstractDenseMatrix {
+
+ /**
+ * If the matrix is upper triangular
+ */
+ UpLo uplo;
+
+ /**
+ * If the matrix is unit diagonal or not unit
+ */
+ Diag diag;
+
+ /**
+ * Leading dimension of the matrix
+ */
+ int ld;
+
+ /**
+ * Constructor for AbstractTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ AbstractTriangDenseMatrix(int n, UpLo uplo, Diag diag) {
+ super(n, n);
+ ld = n;
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ /**
+ * Constructor for AbstractTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ */
+ AbstractTriangDenseMatrix(Matrix A, UpLo uplo, Diag diag) {
+ this(A, Math.min(A.numRows(), A.numColumns()), uplo, diag);
+ }
+
+ /**
+ * Constructor for AbstractTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ AbstractTriangDenseMatrix(Matrix A, boolean deep, UpLo uplo, Diag diag) {
+ this(A, Math.min(A.numRows(), A.numColumns()), deep, uplo, diag);
+ }
+
+ /**
+ * Constructor for AbstractTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ */
+ AbstractTriangDenseMatrix(Matrix A, int k, UpLo uplo, Diag diag) {
+ this(A, k, true, uplo, diag);
+ }
+
+ /**
+ * Constructor for AbstractTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ AbstractTriangDenseMatrix(Matrix A, int k, boolean deep, UpLo uplo,
+ Diag diag) {
+ super(A, deep);
+
+ if (deep && !A.isSquare())
+ throw new IllegalArgumentException("deep && !A.isSquare()");
+
+ ld = A.numRows();
+ numRows = numColumns = k;
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ @Override
+ public Vector mult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.mult(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A*z
+ BLAS.getInstance().dtrmv(uplo.netlib(), Transpose.NoTranspose.netlib(), diag.netlib(),
+ numRows, data, Math.max(1, ld), yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.transMult(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A'*y
+ BLAS.getInstance().dtrmv(uplo.netlib(), Transpose.Transpose.netlib(), diag.netlib(),
+ numRows, data, Math.max(1, ld), yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Matrix mult(double alpha, Matrix B, Matrix C) {
+ if (!(C instanceof DenseMatrix))
+ return super.mult(alpha, B, C);
+
+ checkMultAdd(B, C);
+
+ double[] Cd = ((DenseMatrix) C).getData();
+
+ C.set(B);
+
+ // C = alpha*A*C
+ BLAS.getInstance().dtrmm(Side.Left.netlib(), uplo.netlib(), Transpose.NoTranspose.netlib(),
+ diag.netlib(), C.numRows(), C.numColumns(), alpha, data, Math.max(1, ld), Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix transAmult(double alpha, Matrix B, Matrix C) {
+ if (!(C instanceof DenseMatrix))
+ return super.transAmult(alpha, B, C);
+
+ checkTransAmultAdd(B, C);
+
+ double[] Cd = ((DenseMatrix) C).getData();
+
+ C.set(B);
+
+ // C = alpha*A'*C
+ BLAS.getInstance().dtrmm(Side.Left.netlib(), uplo.netlib(), Transpose.Transpose.netlib(),
+ diag.netlib(), C.numRows(), C.numColumns(), alpha, data, Math.max(1, ld), Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.NoTranspose);
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.Transpose);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ transSolve(B, X);
+ return x;
+ }
+
+ Matrix solve(Matrix B, Matrix X, Transpose trans) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ // Different argument checking to support Hessenberg type matrices for
+ // solvers such as GMRES
+ if (B.numRows() < numRows)
+ throw new IllegalArgumentException("B.numRows() < A.numRows()");
+ if (B.numColumns() != X.numColumns())
+ throw new IllegalArgumentException(
+ "B.numColumns() != X.numColumns()");
+ if (X.numRows() < numRows)
+ throw new IllegalArgumentException("X.numRows() < A.numRows()");
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dtrtrs(uplo.netlib(), trans.netlib(), diag.netlib(), numRows,
+ X.numColumns(), data, Math.max(1, ld), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + column * Math.max(ld, numRows);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new TriangDenseMatrixIterator();
+ }
+
+ private class TriangDenseMatrixIterator extends RefMatrixIterator {
+
+ @Override
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ if (uplo == UpLo.Lower)
+ if (row < numRows - 1)
+ row++;
+ else {
+ column++;
+ row = column;
+ }
+ else { // uplo == UpLo.Upper
+ if (row < column)
+ row++;
+ else {
+ column++;
+ row = 0;
+ }
+ }
+
+ return entry;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractTriangPackMatrix.java b/src/main/java/no/uib/cipr/matrix/AbstractTriangPackMatrix.java
new file mode 100644
index 0000000..c04a139
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractTriangPackMatrix.java
@@ -0,0 +1,186 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+import java.util.Iterator;
+
+
+/**
+ * Partial implementation of a triangular, packed matrix
+ */
+abstract class AbstractTriangPackMatrix extends AbstractPackMatrix {
+
+ /**
+ * Upper or lower triangular
+ */
+ UpLo uplo;
+
+ /**
+ * If the matrix is unit diagonal or not unit
+ */
+ Diag diag;
+
+ /**
+ * Constructor for AbstractTriangPackMatrix
+ */
+ AbstractTriangPackMatrix(int n, UpLo uplo, Diag diag) {
+ super(n);
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ /**
+ * Constructor for AbstractTriangPackMatrix
+ */
+ AbstractTriangPackMatrix(Matrix A, UpLo uplo, Diag diag) {
+ this(A, false, uplo, diag);
+ }
+
+ /**
+ * Constructor for AbstractTriangPackMatrix
+ */
+ AbstractTriangPackMatrix(Matrix A, boolean deep, UpLo uplo, Diag diag) {
+ super(A, deep);
+ this.uplo = uplo;
+ this.diag = diag;
+ }
+
+ @Override
+ public Vector mult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.mult(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A*z
+ BLAS.getInstance().dtpmv(uplo.netlib(), Transpose.NoTranspose.netlib(), diag.netlib(),
+ numRows, data, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMult(double alpha, Vector x, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.transMult(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = alpha*x
+ y.set(alpha, x);
+
+ // y = A*z
+ BLAS.getInstance().dtpmv(uplo.netlib(), Transpose.Transpose.netlib(), diag.netlib(),
+ numRows, data, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.NoTranspose);
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X, Transpose.Transpose);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ transSolve(B, X);
+ return x;
+ }
+
+ Matrix solve(Matrix B, Matrix X, Transpose trans) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dtptrs(uplo.netlib(), trans.netlib(), diag.netlib(), numRows,
+ X.numColumns(), data, Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new TriangPackMatrixIterator();
+ }
+
+ private class TriangPackMatrixIterator extends RefMatrixIterator {
+
+ @Override
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ if (uplo == UpLo.Lower)
+ if (row < numRows - 1)
+ row++;
+ else {
+ column++;
+ row = column;
+ }
+ else { // uplo == UpLo.Upper
+ if (row < column)
+ row++;
+ else {
+ column++;
+ row = 0;
+ }
+ }
+
+ return entry;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/AbstractVector.java b/src/main/java/no/uib/cipr/matrix/AbstractVector.java
new file mode 100644
index 0000000..c210ce2
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/AbstractVector.java
@@ -0,0 +1,304 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.io.Serializable;
+import java.util.Formatter;
+import java.util.Iterator;
+
+/**
+ * Partial implementation of <code>Vector</code>. The following methods throw
+ * <code>UnsupportedOperationException</code>, and should be overridden by a
+ * subclass:
+ * <ul>
+ * <li><code>get(int)</code></li>
+ * <li><code>set(int,double)</code></li>
+ * <li><code>copy</code></li>
+ * </ul>
+ * <p>
+ * For the rest of the methods, simple default implementations using a vector
+ * iterator has been provided. There are some kernel operations which the
+ * simpler operations forward to, and they are:
+ * <ul>
+ * <li> <code>add(double,Vector)</code> and <code>set(double,Vector)</code>.
+ * </li>
+ * <li> <code>scale(double)</code>.</li>
+ * <li><code>dot(Vector)</code> and all the norms. </li>
+ * </ul>
+ * <p>
+ * Finally, a default iterator is provided by this class, which works by calling
+ * the <code>get</code> function. A tailored replacement should be used by
+ * subclasses.
+ * </ul>
+ */
+public abstract class AbstractVector implements Vector, Serializable {
+
+ /**
+ * Size of the vector
+ */
+ protected int size;
+
+ /**
+ * Constructor for AbstractVector.
+ *
+ * @param size
+ * Size of the vector
+ */
+ protected AbstractVector(int size) {
+ if (size < 0)
+ throw new IllegalArgumentException("Vector size cannot be negative");
+ this.size = size;
+ }
+
+ /**
+ * Constructor for AbstractVector, same size as x
+ *
+ * @param x
+ * Vector to get the size from
+ */
+ protected AbstractVector(Vector x) {
+ this.size = x.size();
+ }
+
+ public int size() {
+ return size;
+ }
+
+ public void set(int index, double value) {
+ throw new UnsupportedOperationException();
+ }
+
+ public void add(int index, double value) {
+ set(index, value + get(index));
+ }
+
+ public double get(int index) {
+ throw new UnsupportedOperationException();
+ }
+
+ public Vector copy() {
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Checks the index
+ */
+ protected void check(int index) {
+ if (index < 0)
+ throw new IndexOutOfBoundsException("index is negative (" + index
+ + ")");
+ if (index >= size)
+ throw new IndexOutOfBoundsException("index >= size (" + index
+ + " >= " + size + ")");
+ }
+
+ public Vector zero() {
+ for (VectorEntry e : this)
+ e.set(0);
+ return this;
+ }
+
+ public Vector scale(double alpha) {
+ if (alpha == 0)
+ return zero();
+ else if (alpha == 1)
+ return this;
+
+ for (VectorEntry e : this)
+ e.set(alpha * e.get());
+
+ return this;
+ }
+
+ public Vector set(Vector y) {
+ return set(1, y);
+ }
+
+ public Vector set(double alpha, Vector y) {
+ checkSize(y);
+
+ if (alpha == 0)
+ return zero();
+
+ zero();
+ for (VectorEntry e : y)
+ set(e.index(), alpha * e.get());
+
+ return this;
+ }
+
+ public Vector add(Vector y) {
+ return add(1, y);
+ }
+
+ public Vector add(double alpha, Vector y) {
+ checkSize(y);
+
+ if (alpha == 0)
+ return this;
+
+ for (VectorEntry e : y)
+ add(e.index(), alpha * e.get());
+
+ return this;
+ }
+
+ public double dot(Vector y) {
+ checkSize(y);
+
+ double ret = 0;
+ for (VectorEntry e : this)
+ ret += e.get() * y.get(e.index());
+ return ret;
+ }
+
+ /**
+ * Checks for conformant sizes
+ */
+ protected void checkSize(Vector y) {
+ if (size != y.size())
+ throw new IndexOutOfBoundsException("x.size != y.size (" + size
+ + " != " + y.size() + ")");
+ }
+
+ public double norm(Norm type) {
+ if (type == Norm.One)
+ return norm1();
+ else if (type == Norm.Two)
+ return norm2();
+ else if (type == Norm.TwoRobust)
+ return norm2_robust();
+ else
+ // Infinity
+ return normInf();
+ }
+
+ protected double norm1() {
+ double sum = 0;
+ for (VectorEntry e : this)
+ sum += Math.abs(e.get());
+ return sum;
+ }
+
+ protected double norm2() {
+ double norm = 0;
+ for (VectorEntry e : this)
+ norm += e.get() * e.get();
+ return Math.sqrt(norm);
+ }
+
+ protected double norm2_robust() {
+ double scale = 0, ssq = 1;
+ for (VectorEntry e : this) {
+ double xval = e.get();
+ if (xval != 0) {
+ double absxi = Math.abs(xval);
+ if (scale < absxi) {
+ ssq = 1 + ssq * Math.pow(scale / absxi, 2);
+ scale = absxi;
+ } else
+ ssq = ssq + Math.pow(absxi / scale, 2);
+ }
+ }
+ return scale * Math.sqrt(ssq);
+ }
+
+ protected double normInf() {
+ double max = 0;
+ for (VectorEntry e : this)
+ max = Math.max(Math.abs(e.get()), max);
+ return max;
+ }
+
+ public Iterator<VectorEntry> iterator() {
+ return new RefVectorIterator();
+ }
+
+ @Override
+ public String toString() {
+ // Output into coordinate format. Indices start from 1 instead of 0
+ Formatter out = new Formatter();
+
+ out.format("%10d %19d\n", size, Matrices.cardinality(this));
+
+ for (VectorEntry e : this)
+ if (e.get() != 0)
+ out.format("%10d % .12e\n", e.index() + 1, e.get());
+
+ return out.toString();
+ }
+
+ /**
+ * Iterator over a general vector
+ */
+ private class RefVectorIterator implements Iterator<VectorEntry> {
+
+ private int index;
+
+ private final RefVectorEntry entry = new RefVectorEntry();
+
+ public boolean hasNext() {
+ return index < size;
+ }
+
+ public VectorEntry next() {
+ entry.update(index);
+
+ index++;
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Vector entry backed by the vector. May be reused for higher performance
+ */
+ private class RefVectorEntry implements VectorEntry {
+
+ private int index;
+
+ /**
+ * Updates the entry
+ */
+ public void update(int index) {
+ this.index = index;
+ }
+
+ public int index() {
+ return index;
+ }
+
+ public double get() {
+ return AbstractVector.this.get(index);
+ }
+
+ public void set(double value) {
+ AbstractVector.this.set(index, value);
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/BandCholesky.java b/src/main/java/no/uib/cipr/matrix/BandCholesky.java
new file mode 100644
index 0000000..b782e0e
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/BandCholesky.java
@@ -0,0 +1,254 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrix.Norm;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+/**
+ * Banded Cholesky decomposition
+ */
+public class BandCholesky {
+
+ /**
+ * Matrix dimension
+ */
+ private final int n;
+
+ /**
+ * Number of bands in the matrix A
+ */
+ private final int kd;
+
+ /**
+ * Cholesky decomposition of a lower matrix
+ */
+ private LowerTriangBandMatrix Cl;
+
+ /**
+ * Cholesky decomposition of an upper matrix
+ */
+ private UpperTriangBandMatrix Cu;
+
+ /**
+ * If the matrix is SPD or not
+ */
+ private boolean notspd;
+
+ /**
+ * True for upper part, else false
+ */
+ private final boolean upper;
+
+ /**
+ * Constructor for BandCholesky
+ *
+ * @param n
+ * Matrix size
+ * @param kd
+ * Number of matrix bands
+ * @param upper
+ * True for decomposing an upper symmetrical matrix, false for a
+ * lower symmetrical matrix
+ */
+ public BandCholesky(int n, int kd, boolean upper) {
+ this.n = n;
+ this.kd = kd;
+ this.upper = upper;
+
+ if (upper)
+ Cu = new UpperTriangBandMatrix(n, kd);
+ else
+ Cl = new LowerTriangBandMatrix(n, kd);
+ }
+
+ /**
+ * Creates a Cholesky decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return A Cholesky decomposition of the matrix
+ */
+ public static BandCholesky factorize(LowerSPDBandMatrix A) {
+ return new BandCholesky(A.numRows(), A.kl, false).factor(A);
+ }
+
+ /**
+ * Creates a Cholesky decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return A Cholesky decomposition of the matrix
+ */
+ public static BandCholesky factorize(UpperSPDBandMatrix A) {
+ return new BandCholesky(A.numRows(), A.ku, true).factor(A);
+ }
+
+ /**
+ * Creates a Cholesky decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public BandCholesky factor(LowerSPDBandMatrix A) {
+ if (upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for upper matrices");
+
+ return decompose(A);
+ }
+
+ /**
+ * Creates a Cholesky decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public BandCholesky factor(UpperSPDBandMatrix A) {
+ if (!upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for lower matrices");
+
+ return decompose(A);
+ }
+
+ private BandCholesky decompose(AbstractBandMatrix A) {
+ if (n != A.numRows())
+ throw new IllegalArgumentException("n != A.numRows()");
+ if (upper && A.ku != kd)
+ throw new IllegalArgumentException("A.ku != kd");
+ if (!upper && A.kl != kd)
+ throw new IllegalArgumentException("A.kl != kd");
+
+ notspd = false;
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpbtrf(UpLo.Upper.netlib(), n, kd, A.getData(),
+ Matrices.ld(kd + 1), info);
+ else
+ LAPACK.getInstance().dpbtrf(UpLo.Lower.netlib(), n, kd, A.getData(),
+ Matrices.ld(kd + 1), info);
+
+ if (info.val > 0)
+ notspd = true;
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ if (upper)
+ Cu.set(A);
+ else
+ Cl.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a lower
+ * SPD matrix
+ */
+ public LowerTriangBandMatrix getL() {
+ if (!upper)
+ return Cl;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a upper
+ * SPD matrix
+ */
+ public UpperTriangBandMatrix getU() {
+ if (upper)
+ return Cu;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Returns true if the matrix decomposed is symmetrical, positive definite
+ */
+ public boolean isSPD() {
+ return !notspd;
+ }
+
+ /**
+ * Computes the reciprocal condition number
+ *
+ * @param A
+ * The matrix this is a decomposition of
+ * @return The reciprocal condition number. Values close to unity indicate a
+ * well-conditioned system, while numbers close to zero do not.
+ */
+ public double rcond(Matrix A) {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+
+ double anorm = A.norm(Norm.One);
+
+ double[] work = new double[3 * n];
+ int[] lwork = new int[n];
+
+ intW info = new intW(0);
+ doubleW rcond = new doubleW(0);
+ if (upper)
+ LAPACK.getInstance().dpbcon(UpLo.Upper.netlib(), n, kd, Cu.getData(),
+ Matrices.ld(kd + 1), anorm, rcond, work, lwork, info);
+ else
+ LAPACK.getInstance().dpbcon(UpLo.Lower.netlib(), n, kd, Cl.getData(),
+ Matrices.ld(kd + 1), anorm, rcond, work, lwork, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return rcond.val;
+ }
+
+ /**
+ * Computes <code>A\B</code>, overwriting <code>B</code>
+ */
+ public DenseMatrix solve(DenseMatrix B) throws MatrixNotSPDException {
+ if (notspd)
+ throw new MatrixNotSPDException();
+ if (B.numRows() != n)
+ throw new IllegalArgumentException("B.numRows() != n");
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpbtrs(UpLo.Upper.netlib(), n, kd, B.numColumns(),
+ Cu.getData(), Matrices.ld(kd + 1), B.getData(), Matrices.ld(n), info);
+ else
+ LAPACK.getInstance().dpbtrs(UpLo.Lower.netlib(), n, kd, B.numColumns(),
+ Cl.getData(), Matrices.ld(kd + 1), B.getData(), Matrices.ld(n), info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return B;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/BandLU.java b/src/main/java/no/uib/cipr/matrix/BandLU.java
new file mode 100644
index 0000000..ea542ba
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/BandLU.java
@@ -0,0 +1,241 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrix.Norm;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+/**
+ * Banded LU decomposition
+ */
+public class BandLU {
+
+ /**
+ * Matrix dimension
+ */
+ private final int n;
+
+ /**
+ * Number of bands in the matrix A
+ */
+ private final int kl, ku;
+
+ /**
+ * Holds the LU factors
+ */
+ private final BandMatrix LU;
+
+ /**
+ * Row pivotations
+ */
+ private final int[] ipiv;
+
+ /**
+ * True if the matrix was singular
+ */
+ private boolean singular;
+
+ /**
+ * Constructor for BandLU
+ *
+ * @param n
+ * Matrix size
+ * @param kl
+ * Number of lower matrix bands
+ * @param ku
+ * Number of upper matrix bands
+ */
+ public BandLU(int n, int kl, int ku) {
+ this.n = n;
+ this.kl = kl;
+ this.ku = ku;
+
+ LU = new BandMatrix(n, kl, ku + kl);
+
+ ipiv = new int[n];
+ }
+
+ /**
+ * Creates an LU decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return A LU decomposition of the matrix
+ */
+ public static BandLU factorize(BandMatrix A) {
+ return new BandLU(A.numRows(), A.kl, A.ku).factor(A, false);
+ }
+
+ /**
+ * Creates an LU decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. If the decomposition is in-place, its
+ * number of superdiagonals must equal <code>kl+ku</code>
+ * @param inplace
+ * Wheter or not the decomposition should overwrite the passed
+ * matrix
+ * @return The current decomposition
+ */
+ public BandLU factor(BandMatrix A, boolean inplace) {
+ if (inplace)
+ return factor(A);
+ else
+ return factor(new BandMatrix(A, kl, kl + ku));
+ }
+
+ /**
+ * Creates an LU decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. It will be overwritten with the
+ * decomposition. Its number of superdiagonals must equal
+ * <code>kl+ku</code>
+ * @return The current decomposition
+ */
+ public BandLU factor(BandMatrix A) {
+ if (!(A.isSquare()))
+ throw new IllegalArgumentException("!A.isSquare()");
+ if (n != A.numRows())
+ throw new IllegalArgumentException("n != A.numRows()");
+ if (A.ku != ku + kl)
+ throw new IllegalArgumentException("A.ku != ku + kl");
+
+ singular = false;
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgbtrf(n, n, kl, ku, A.getData(), 2 * kl + ku + 1, ipiv, info);
+
+ if (info.val > 0)
+ singular = true;
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ LU.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the lower triangular factor
+ */
+ public UnitLowerTriangBandMatrix getL() {
+ return new UnitLowerTriangBandMatrix(LU, LU.numSubDiagonals(), false);
+ }
+
+ /**
+ * Returns the upper triangular factor
+ */
+ public UpperTriangBandMatrix getU() {
+ return new UpperTriangBandMatrix(LU, LU.numSuperDiagonals(), false);
+ }
+
+ /**
+ * Returns the decomposition matrix
+ */
+ public BandMatrix getLU() {
+ return LU;
+ }
+
+ /**
+ * Returns the row pivots
+ */
+ public int[] getPivots() {
+ return ipiv;
+ }
+
+ /**
+ * Checks for singularity
+ */
+ public boolean isSingular() {
+ return singular;
+ }
+
+ /**
+ * Computes the reciprocal condition number, using either the infinity norm
+ * of the 1 norm.
+ *
+ * @param A
+ * The matrix this is a decomposition of
+ * @param norm
+ * Either <code>Norm.One</code> or <code>Norm.Infinity</code>
+ * @return The reciprocal condition number. Values close to unity indicate a
+ * well-conditioned system, while numbers close to zero do not.
+ */
+ public double rcond(Matrix A, Norm norm) {
+ if (norm != Norm.One && norm != Norm.Infinity)
+ throw new IllegalArgumentException(
+ "Only the 1 or the Infinity norms are supported");
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+
+ double anorm = A.norm(norm);
+
+ double[] work = new double[3 * n];
+ int[] lwork = new int[n];
+
+ intW info = new intW(0);
+ doubleW rcond = new doubleW(0);
+ LAPACK.getInstance().dgbcon(norm.netlib(), n, kl, ku, LU.getData(),
+ Matrices.ld(2 * kl + ku + 1), ipiv, anorm, rcond, work, lwork, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return rcond.val;
+ }
+
+ /**
+ * Computes <code>A\B</code>, overwriting <code>B</code>
+ */
+ public DenseMatrix solve(DenseMatrix B) throws MatrixSingularException {
+ return solve(B, Transpose.NoTranspose);
+ }
+
+ /**
+ * Computes <code>A<sup>T</sup>\B</code>, overwriting <code>B</code>
+ */
+ public DenseMatrix transSolve(DenseMatrix B) throws MatrixSingularException {
+ return solve(B, Transpose.Transpose);
+ }
+
+ private DenseMatrix solve(DenseMatrix B, Transpose trans)
+ throws MatrixSingularException {
+ if (singular)
+ throw new MatrixSingularException();
+ if (B.numRows() != n)
+ throw new IllegalArgumentException("B.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgbtrs(trans.netlib(), n, kl, ku, B.numColumns(),
+ LU.getData(), 2 * kl + ku + 1, ipiv, B.getData(), Matrices.ld(n), info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return B;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/BandMatrix.java b/src/main/java/no/uib/cipr/matrix/BandMatrix.java
new file mode 100644
index 0000000..3ef5606
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/BandMatrix.java
@@ -0,0 +1,255 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Banded matrix. The banded matrix is a useful sparse structure for many kinds
+ * of direct computations, however it should only be used if the band is
+ * sufficiently narrow as wide bands actually wastes both memory and compute
+ * time. The matrix
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>12</sub></td>
+ * <td> </td>
+ * <td> </td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td> </td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>34</sub></td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td> </td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>44</sub></td>
+ * <td>a<sub>45</sub></td>
+ * </tr>
+ * <tr>
+ * <td> </td>
+ * <td> </td>
+ * <td>a<sub>53</sub></td>
+ * <td>a<sub>54</sub></td>
+ * <td>a<sub>55</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ * <p>
+ * has two lower diagonals and one upper diagonal. It will be stored in the
+ * array
+ * </p>
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td> </td>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>53</sub></td>
+ * <td>a<sub>34</sub></td>
+ * <td>a<sub>44</sub></td>
+ * <td>a<sub>54</sub></td>
+ * <td> </td>
+ * <td>a<sub>45</sub></td>
+ * <td>a<sub>55</sub></td>
+ * <td> </td>
+ * <td> </td>
+ * </tr>
+ * </table>
+ * </p>
+ * <p>
+ * Empty cells are allocated, but never referenced.
+ * </p>
+ */
+public class BandMatrix extends AbstractBandMatrix {
+
+ /**
+ * Constructor for BandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kl
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param ku
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public BandMatrix(int n, int kl, int ku) {
+ super(n, kl, ku);
+ }
+
+ /**
+ * Constructor for BandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kl
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param ku
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public BandMatrix(Matrix A, int kl, int ku) {
+ super(A, kl, ku);
+ }
+
+ /**
+ * Constructor for BandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kl
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param ku
+ * Number of bands below the main diagonal (subdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public BandMatrix(Matrix A, int kl, int ku, boolean deep) {
+ super(A, kl, ku, deep);
+ }
+
+ @Override
+ public BandMatrix copy() {
+ return new BandMatrix(this, kl, ku);
+ }
+
+ @Override
+ public Matrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dgbmv(Transpose.NoTranspose.netlib(), numRows, numColumns, kl,
+ ku, alpha, data, kl + ku + 1, xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dgbmv(Transpose.Transpose.netlib(), numRows, numColumns, kl, ku,
+ alpha, data, kl + ku + 1, xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ // Allocate factorization matrix. The factorization matrix will be
+ // large enough to accomodate any pivots
+ BandMatrix Af = new BandMatrix(this, kl, ku + kl);
+ int[] ipiv = new int[numRows];
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgbsv(numRows, kl, ku, X.numColumns(),
+ Af.getData(), Matrices.ld(2 * kl + ku + 1), ipiv, Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transpose() {
+ checkTranspose();
+
+ if (kl != ku)
+ throw new IllegalArgumentException("kl != ku");
+
+ for (int j = 0; j < numColumns; ++j)
+ for (int i = j + 1; i < Math.min(j + kl + 1, numRows); ++i) {
+ double value = get(i, j);
+ set(i, j, get(j, i));
+ set(j, i, value);
+ }
+
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/DenseCholesky.java b/src/main/java/no/uib/cipr/matrix/DenseCholesky.java
new file mode 100644
index 0000000..ac67328
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/DenseCholesky.java
@@ -0,0 +1,235 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrix.Norm;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+/**
+ * Dense Cholesky decomposition
+ */
+public class DenseCholesky {
+
+ /**
+ * Matrix dimension
+ */
+ private final int n;
+
+ /**
+ * Cholesky decomposition of a lower matrix
+ */
+ private LowerTriangDenseMatrix Cl;
+
+ /**
+ * Cholesky decomposition of an upper matrix
+ */
+ private UpperTriangDenseMatrix Cu;
+
+ /**
+ * If the matrix is SPD or not
+ */
+ private boolean notspd;
+
+ /**
+ * True for upper part, else false
+ */
+ private final boolean upper;
+
+ /**
+ * Constructor for DenseCholesky
+ *
+ * @param n
+ * Matrix size
+ * @param upper
+ * True for decomposing an upper symmetrical matrix, false for a
+ * lower symmetrical matrix
+ */
+ public DenseCholesky(int n, boolean upper) {
+ this.n = n;
+ this.upper = upper;
+
+ if (upper)
+ Cu = new UpperTriangDenseMatrix(n);
+ else
+ Cl = new LowerTriangDenseMatrix(n);
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return The current decomposition
+ */
+ public static DenseCholesky factorize(Matrix A) {
+ return new DenseCholesky(A.numRows(), true)
+ .factor(new UpperSPDDenseMatrix(A));
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public DenseCholesky factor(LowerSPDDenseMatrix A) {
+ if (upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for upper matrices");
+
+ return decompose(A);
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public DenseCholesky factor(UpperSPDDenseMatrix A) {
+ if (!upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for lower matrices");
+
+ return decompose(A);
+ }
+
+ private DenseCholesky decompose(AbstractDenseMatrix A) {
+ if (n != A.numRows())
+ throw new IllegalArgumentException("n != A.numRows()");
+
+ notspd = false;
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpotrf(UpLo.Upper.netlib(), A.numRows(),
+ A.getData(), Matrices.ld(A.numRows()), info);
+ else
+ LAPACK.getInstance().dpotrf(UpLo.Lower.netlib(), A.numRows(),
+ A.getData(), Matrices.ld(A.numRows()), info);
+
+ if (info.val > 0)
+ notspd = true;
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ if (upper)
+ Cu.set(A);
+ else
+ Cl.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns true if the matrix decomposed is symmetrical, positive definite
+ */
+ public boolean isSPD() {
+ return !notspd;
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a lower
+ * SPD matrix
+ */
+ public LowerTriangDenseMatrix getL() {
+ if (!upper)
+ return Cl;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a upper
+ * SPD matrix
+ */
+ public UpperTriangDenseMatrix getU() {
+ if (upper)
+ return Cu;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Solves for <code>B</code>, overwriting it on return
+ */
+ public DenseMatrix solve(DenseMatrix B) throws MatrixNotSPDException {
+ if (notspd)
+ throw new MatrixNotSPDException();
+ if (n != B.numRows())
+ throw new IllegalArgumentException("n != B.numRows()");
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpotrs(UpLo.Upper.netlib(), Cu.numRows(),
+ B.numColumns(), Cu.getData(), Matrices.ld(Cu.numRows()),
+ B.getData(), Matrices.ld(Cu.numRows()), info);
+ else
+ LAPACK.getInstance().dpotrs(UpLo.Lower.netlib(), Cl.numRows(),
+ B.numColumns(), Cl.getData(), Matrices.ld(Cl.numRows()),
+ B.getData(), Matrices.ld(Cl.numRows()), info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return B;
+ }
+
+ /**
+ * Computes the reciprocal condition number
+ *
+ * @param A
+ * The matrix this is a decomposition of
+ * @return The reciprocal condition number. Values close to unity indicate a
+ * well-conditioned system, while numbers close to zero do not.
+ */
+ public double rcond(Matrix A) {
+ if (n != A.numRows())
+ throw new IllegalArgumentException("n != A.numRows()");
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+
+ double anorm = A.norm(Norm.One);
+
+ double[] work = new double[3 * n];
+ int[] iwork = new int[n];
+
+ intW info = new intW(0);
+ doubleW rcond = new doubleW(0);
+ if (upper)
+ LAPACK.getInstance().dpocon(UpLo.Upper.netlib(), n, Cu.getData(),
+ Matrices.ld(n), anorm, rcond, work, iwork, info);
+ else
+ LAPACK.getInstance().dpocon(UpLo.Lower.netlib(), n, Cl.getData(),
+ Matrices.ld(n), anorm, rcond, work, iwork, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return rcond.val;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/DenseLU.java b/src/main/java/no/uib/cipr/matrix/DenseLU.java
new file mode 100644
index 0000000..207f537
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/DenseLU.java
@@ -0,0 +1,203 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import no.uib.cipr.matrix.Matrix.Norm;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+/**
+ * Dense Partial Pivot LU decomposition: {@code A = P * L * U}.
+ */
+public class DenseLU {
+
+ /**
+ * Holds the LU factors
+ */
+ private DenseMatrix LU;
+
+ /**
+ * Row pivotations
+ */
+ private int[] piv;
+
+ /**
+ * True if the matrix was singular
+ */
+ private boolean singular;
+
+ /**
+ * Constructor for DenseLU
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns
+ */
+ public DenseLU(int m, int n) {
+ LU = new DenseMatrix(m, n);
+ piv = new int[Math.min(m, n)];
+ }
+
+ /**
+ * Creates an LU decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return The current decomposition
+ */
+ public static DenseLU factorize(Matrix A) {
+ return new DenseLU(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ /**
+ * Creates an LU decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to decompose. Overwritten with the decomposition
+ * @return The current decomposition
+ */
+ public DenseLU factor(DenseMatrix A) {
+ singular = false;
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgetrf(A.numRows(), A.numColumns(),
+ A.getData(), Matrices.ld(A.numRows()), piv, info);
+
+ if (info.val > 0)
+ singular = true;
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ LU.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the permutation matrix.
+ */
+ public PermutationMatrix getP() {
+ PermutationMatrix perm = PermutationMatrix.fromPartialPivots(piv);
+ perm.transpose();
+ return perm;
+ }
+
+ /**
+ * Returns the lower triangular factor
+ */
+ public UnitLowerTriangDenseMatrix getL() {
+ return new UnitLowerTriangDenseMatrix(getLU(), false);
+ }
+
+ /**
+ * Returns the upper triangular factor
+ */
+ public UpperTriangDenseMatrix getU() {
+ return new UpperTriangDenseMatrix(getLU(), false);
+ }
+
+ /**
+ * Returns the decomposition matrix
+ */
+ protected DenseMatrix getLU() {
+ return LU;
+ }
+
+ /**
+ * Computes the reciprocal condition number, using either the infinity norm
+ * of the 1 norm.
+ *
+ * @param A
+ * The matrix this is a decomposition of
+ * @param norm
+ * Either <code>Norm.One</code> or <code>Norm.Infinity</code>
+ * @return The reciprocal condition number. Values close to unity indicate a
+ * well-conditioned system, while numbers close to zero do not.
+ */
+ public double rcond(Matrix A, Norm norm) {
+ if (norm != Norm.One && norm != Norm.Infinity)
+ throw new IllegalArgumentException(
+ "Only the 1 or the Infinity norms are supported");
+
+ double anorm = A.norm(norm);
+
+ int n = A.numRows();
+
+ intW info = new intW(0);
+ doubleW rcond = new doubleW(0);
+ LAPACK.getInstance().dgecon(norm.netlib(), n, LU.getData(), Matrices.ld(n), anorm,
+ rcond, new double[4 * n], new int[n], info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return rcond.val;
+ }
+
+ /**
+ * Returns the row pivots
+ */
+ public int[] getPivots() {
+ return piv;
+ }
+
+ /**
+ * Checks for singularity
+ */
+ public boolean isSingular() {
+ return singular;
+ }
+
+ /**
+ * Computes <code>A\B</code>, overwriting <code>B</code>
+ */
+ public DenseMatrix solve(DenseMatrix B) throws MatrixSingularException {
+ return solve(B, Transpose.NoTranspose);
+ }
+
+ /**
+ * Computes <code>A<sup>T</sup>\B</code>, overwriting <code>B</code>
+ */
+ public DenseMatrix transSolve(DenseMatrix B) throws MatrixSingularException {
+ return solve(B, Transpose.Transpose);
+ }
+
+ private DenseMatrix solve(DenseMatrix B, Transpose trans)
+ throws MatrixSingularException {
+ if (singular)
+ throw new MatrixSingularException();
+ if (B.numRows() != LU.numRows())
+ throw new IllegalArgumentException("B.numRows() != LU.numRows()");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgetrs(trans.netlib(), LU.numRows(),
+ B.numColumns(), LU.getData(), Matrices.ld(LU.numRows()),
+ piv, B.getData(), Matrices.ld(LU.numRows()), info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return B;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/DenseMatrix.java b/src/main/java/no/uib/cipr/matrix/DenseMatrix.java
new file mode 100644
index 0000000..5aca14e
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/DenseMatrix.java
@@ -0,0 +1,517 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.io.IOException;
+
+import no.uib.cipr.matrix.io.MatrixInfo;
+import no.uib.cipr.matrix.io.MatrixSize;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+import com.github.fommil.netlib.BLAS;
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Dense matrix. It is a good all-round matrix structure, with fast access and
+ * efficient algebraic operations. The matrix
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>12</sub></td>
+ * <td>a<sub>13</sub></td>
+ * <td>a<sub>14</sub></td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td>a<sub>24</sub></td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>34</sub></td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>41</sub></td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ * <p>
+ * is stored column major in a single array, as follows:
+ * </p>
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>41</sub></td>
+ * <td>a<sub>12</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>13</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>14</sub></td>
+ * <td>a<sub>24</sub></td>
+ * <td>a<sub>34</sub></td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ */
+public class DenseMatrix extends AbstractDenseMatrix {
+
+ /**
+ * Constructor for DenseMatrix
+ *
+ * @param r
+ * Reader to get the matrix from
+ */
+ public DenseMatrix(MatrixVectorReader r) throws IOException {
+ // Start with a zero-sized matrix
+ super(0, 0);
+
+ // Get matrix information. Use the header if present, else use a safe
+ // default
+ MatrixInfo info = null;
+ if (r.hasInfo())
+ info = r.readMatrixInfo();
+ else
+ info = new MatrixInfo(true, MatrixInfo.MatrixField.Real,
+ MatrixInfo.MatrixSymmetry.General);
+ MatrixSize size = r.readMatrixSize(info);
+
+ // Resize the matrix to correct size
+ numRows = size.numRows();
+ numColumns = size.numColumns();
+ data = new double[numRows * numColumns];
+
+ // Check that the matrix is in an acceptable format
+ if (info.isPattern())
+ throw new UnsupportedOperationException(
+ "Pattern matrices are not supported");
+ if (info.isComplex())
+ throw new UnsupportedOperationException(
+ "Complex matrices are not supported");
+
+ // Read the entries, in either coordinate or array format
+ if (info.isCoordinate()) {
+
+ // Read coordinate data
+ int nz = size.numEntries();
+ int[] row = new int[nz];
+ int[] column = new int[nz];
+ double[] entry = new double[nz];
+ r.readCoordinate(row, column, entry);
+
+ // Shift indices from 1-offset to 0-offset
+ r.add(-1, row);
+ r.add(-1, column);
+
+ // Store them
+ for (int i = 0; i < nz; ++i)
+ set(row[i], column[i], entry[i]);
+
+ } else
+ // info.isArray()
+ r.readArray(data);
+
+ // Put in missing entries from symmetry or skew symmetry
+ if (info.isSymmetric())
+ for (int i = 0; i < numRows; ++i)
+ for (int j = 0; j < i; ++j)
+ set(j, i, get(i, j));
+ else if (info.isSkewSymmetric())
+ for (int i = 0; i < numRows; ++i)
+ for (int j = 0; j < i; ++j)
+ set(j, i, -get(i, j));
+ }
+
+ /**
+ * Constructor for DenseMatrix
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of columns
+ */
+ public DenseMatrix(int numRows, int numColumns) {
+ super(numRows, numColumns);
+ }
+
+ /**
+ * Constructor for DenseMatrix
+ *
+ * @param A
+ * Matrix to copy. A deep copy is made
+ */
+ public DenseMatrix(Matrix A) {
+ super(A);
+ }
+
+ /**
+ * Constructor for DenseMatrix
+ *
+ * @param A
+ * Matrix to copy contents from
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ public DenseMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ /**
+ * Constructor for DenseMatrix. Builds the matrix from a vector
+ *
+ * @param x
+ * Vector to copy from. This will form this matrix' single column
+ * @param deep
+ * If true, x is copied, if false, the internal storage of this
+ * matrix is the same as that of the vector. In that case,
+ * <code>x</code> must be a <code>DenseVector</code>
+ */
+ public DenseMatrix(Vector x, boolean deep) {
+ super(x.size(), 1);
+
+ if (deep)
+ for (VectorEntry e : x)
+ set(e.index(), 0, e.get());
+ else {
+ if (!(x instanceof DenseVector))
+ throw new IllegalArgumentException("x must be a DenseVector");
+ data = ((DenseVector) x).getData();
+ }
+ }
+
+ /**
+ * Constructor for DenseMatrix. Builds the matrix from a vector
+ *
+ * @param x
+ * The vector which forms this matrix' single column. It is
+ * copied, not referenced
+ */
+ public DenseMatrix(Vector x) {
+ this(x, true);
+ }
+
+ /**
+ * Constructor for DenseMatrix. Builds the matrix from vectors. Each vector
+ * will correspond to a column of the matrix
+ *
+ * @param x
+ * Vectors which forms the columns of this matrix. Every vector
+ * must have the same size
+ */
+ public DenseMatrix(Vector[] x) {
+ super(x[0].size(), x.length);
+
+ // Ensure correct sizes
+ for (Vector v : x)
+ if (v.size() != numRows)
+ throw new IllegalArgumentException(
+ "All vectors must be of the same size");
+
+ // Copy the contents
+ for (int j = 0; j < x.length; ++j)
+ for (VectorEntry e : x[j])
+ set(e.index(), j, e.get());
+ }
+
+ /**
+ * Constructor for DenseMatrix. Copies from the passed array
+ *
+ * @param values
+ * Arrays to copy from. Every sub-array must have the same size
+ */
+ public DenseMatrix(double[][] values) {
+ super(values.length, values[0].length);
+
+ // Copy the contents
+ for (int i = 0; i < values.length; ++i) {
+ if (values[i].length != numColumns)
+ throw new IllegalArgumentException("Array cannot be jagged");
+ for (int j = 0; j < values[i].length; ++j)
+ set(i, j, values[i][j]);
+ }
+ }
+
+ @Override
+ public DenseMatrix copy() {
+ return new DenseMatrix(this);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public Matrix multAdd(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.multAdd(alpha, B, C);
+
+ checkMultAdd(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dgemm(Transpose.NoTranspose.netlib(), Transpose.NoTranspose.netlib(),
+ C.numRows(), C.numColumns(), numColumns, alpha, data,
+ Math.max(1, numRows), Bd, Math.max(1, B.numRows()), 1, Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix transAmultAdd(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.transAmultAdd(alpha, B, C);
+
+ checkTransAmultAdd(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dgemm(Transpose.Transpose.netlib(), Transpose.NoTranspose.netlib(),
+ C.numRows(), C.numColumns(), numRows, alpha, data,
+ Math.max(1, numRows), Bd, Math.max(1, B.numRows()), 1, Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix transBmultAdd(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.transBmultAdd(alpha, B, C);
+
+ checkTransBmultAdd(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dgemm(Transpose.NoTranspose.netlib(), Transpose.Transpose.netlib(),
+ C.numRows(), C.numColumns(), numColumns, alpha, data,
+ Math.max(1, numRows), Bd, Math.max(1, B.numRows()), 1, Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix transABmultAdd(double alpha, Matrix B, Matrix C) {
+ if (!(B instanceof DenseMatrix) || !(C instanceof DenseMatrix))
+ return super.transABmultAdd(alpha, B, C);
+
+ checkTransABmultAdd(B, C);
+
+ double[] Bd = ((DenseMatrix) B).getData(), Cd = ((DenseMatrix) C)
+ .getData();
+
+ BLAS.getInstance().dgemm(Transpose.Transpose.netlib(), Transpose.Transpose.netlib(),
+ C.numRows(), C.numColumns(), numRows, alpha, data,
+ Math.max(1, numRows), Bd, Math.max(1, B.numRows()), 1, Cd,
+ Math.max(1, C.numRows()));
+
+ return C;
+ }
+
+ @Override
+ public Matrix rank1(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.rank1(alpha, x, y);
+
+ checkRank1(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dger(numRows, numColumns, alpha, xd, 1, yd, 1, data,
+ Math.max(1, numRows));
+
+ return this;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dgemv(Transpose.NoTranspose.netlib(), numRows, numColumns,
+ alpha, data, Math.max(numRows, 1), xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ BLAS.getInstance().dgemv(Transpose.Transpose.netlib(), numRows, numColumns, alpha,
+ data, Math.max(numRows, 1), xd, 1, 1, yd, 1);
+
+ return y;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ // We allow non-square matrices, as we then use a least-squares solver
+ if (numRows != B.numRows())
+ throw new IllegalArgumentException("numRows != B.numRows() ("
+ + numRows + " != " + B.numRows() + ")");
+ if (numColumns != X.numRows())
+ throw new IllegalArgumentException("numColumns != X.numRows() ("
+ + numColumns + " != " + X.numRows() + ")");
+ if (X.numColumns() != B.numColumns())
+ throw new IllegalArgumentException(
+ "X.numColumns() != B.numColumns() (" + X.numColumns()
+ + " != " + B.numColumns() + ")");
+
+ if (isSquare())
+ return LUsolve(B, X);
+ else
+ return QRsolve(B, X, Transpose.NoTranspose);
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ // We allow non-square matrices, as we then use a least-squares solver
+ if (numColumns != B.numRows())
+ throw new IllegalArgumentException("numColumns != B.numRows() ("
+ + numColumns + " != " + B.numRows() + ")");
+ if (numRows != X.numRows())
+ throw new IllegalArgumentException("numRows != X.numRows() ("
+ + numRows + " != " + X.numRows() + ")");
+ if (X.numColumns() != B.numColumns())
+ throw new IllegalArgumentException(
+ "X.numColumns() != B.numColumns() (" + X.numColumns()
+ + " != " + B.numColumns() + ")");
+
+ return QRsolve(B, X, Transpose.Transpose);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ transSolve(B, X);
+ return x;
+ }
+
+ Matrix LUsolve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ int[] piv = new int[numRows];
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgesv(numRows, B.numColumns(),
+ data.clone(), Matrices.ld(numRows), piv, Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ Matrix QRsolve(Matrix B, Matrix X, Transpose trans) {
+ int nrhs = B.numColumns();
+
+ // Allocate temporary solution matrix
+ DenseMatrix Xtmp = new DenseMatrix(Math.max(numRows, numColumns), nrhs);
+ int M = trans == Transpose.NoTranspose ? numRows : numColumns;
+ for (int j = 0; j < nrhs; ++j)
+ for (int i = 0; i < M; ++i)
+ Xtmp.set(i, j, B.get(i, j));
+ double[] newData = data.clone();
+
+ // Query optimal workspace
+ double[] work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgels(trans.netlib(), numRows, numColumns, nrhs,
+ newData, Matrices.ld(numRows), Xtmp.getData(), Matrices.ld(numRows, numColumns),
+ work, -1, info);
+
+ // Allocate workspace
+ int lwork = -1;
+ if (info.val != 0)
+ lwork = Math.max(1, Math.min(numRows, numColumns)
+ + Math.max(Math.min(numRows, numColumns), nrhs));
+ else
+ lwork = Math.max((int) work[0], 1);
+ work = new double[lwork];
+
+ // Compute the factorization
+ info.val = 0;
+ LAPACK.getInstance().dgels(trans.netlib(), numRows, numColumns, nrhs,
+ newData, Matrices.ld(numRows), Xtmp.getData(), Matrices.ld(numRows, numColumns),
+ work, lwork, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ // Extract the solution
+ int N = trans == Transpose.NoTranspose ? numColumns : numRows;
+ for (int j = 0; j < nrhs; ++j)
+ for (int i = 0; i < N; ++i)
+ X.set(i, j, Xtmp.get(i, j));
+ return X;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/DenseVector.java b/src/main/java/no/uib/cipr/matrix/DenseVector.java
new file mode 100644
index 0000000..d11c227
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/DenseVector.java
@@ -0,0 +1,328 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.io.IOException;
+import java.io.Serializable;
+import java.util.Arrays;
+
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+import no.uib.cipr.matrix.io.VectorInfo;
+import no.uib.cipr.matrix.io.VectorSize;
+import no.uib.cipr.matrix.io.VectorInfo.VectorField;
+
+/**
+ * Dense vector. Stored by a <code>double[]</code> array of the same length as
+ * the vector itself.
+ */
+public class DenseVector extends AbstractVector implements Serializable {
+
+ /** just the private data */
+ private static final long serialVersionUID = 5358813524094629362L;
+
+ /**
+ * Vector data
+ */
+ private final double[] data;
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param r
+ * Reader to get vector from
+ */
+ public DenseVector(MatrixVectorReader r) throws IOException {
+ // Start with a zero-sized vector
+ super(0);
+
+ // Get vector information. Use the header if present, else use a safe
+ // default
+ VectorInfo info = null;
+ if (r.hasInfo())
+ info = r.readVectorInfo();
+ else
+ info = new VectorInfo(true, VectorField.Real);
+ VectorSize size = r.readVectorSize(info);
+
+ // Resize the vector to correct size
+ this.size = size.size();
+ data = new double[size.size()];
+
+ // Check that the vector is in an acceptable format
+ if (info.isPattern())
+ throw new UnsupportedOperationException(
+ "Pattern vectors are not supported");
+ if (info.isComplex())
+ throw new UnsupportedOperationException(
+ "Complex vectors are not supported");
+
+ // Read the entries
+ if (info.isCoordinate()) {
+
+ // Read coordinate data
+ int nz = size.numEntries();
+ int[] index = new int[nz];
+ double[] entry = new double[nz];
+ r.readCoordinate(index, entry);
+
+ // Shift indices from 1-offset to 0-offset
+ r.add(-1, index);
+
+ // Store them
+ for (int i = 0; i < nz; ++i)
+ set(index[i], entry[i]);
+
+ } else
+ // info.isArray()
+ r.readArray(data);
+ }
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param size
+ * Size of the vector
+ */
+ public DenseVector(int size) {
+ super(size);
+ data = new double[size];
+ }
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param x
+ * Copies contents from this vector. A deep copy is made
+ */
+ public DenseVector(Vector x) {
+ this(x, true);
+ }
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param x
+ * Copies contents from this vector
+ * @param deep
+ * True for a deep copy. For a shallow copy, <code>x</code>
+ * must be a <code>DenseVector</code>
+ */
+ public DenseVector(Vector x, boolean deep) {
+ super(x);
+
+ if (deep) {
+ data = new double[size];
+ set(x);
+ } else
+ data = ((DenseVector) x).getData();
+ }
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param x
+ * Copies contents from this array
+ * @param deep
+ * True for a deep copy. For a shallow copy, <code>x</code> is
+ * aliased with the internal storage
+ */
+ public DenseVector(double[] x, boolean deep) {
+ super(x.length);
+
+ if (deep)
+ data = x.clone();
+ else
+ data = x;
+ }
+
+ /**
+ * Constructor for DenseVector
+ *
+ * @param x
+ * Copies contents from this array in a deep copy
+ */
+ public DenseVector(double[] x) {
+ this(x, true);
+ }
+
+ @Override
+ public void set(int index, double value) {
+ check(index);
+ data[index] = value;
+ }
+
+ @Override
+ public void add(int index, double value) {
+ check(index);
+ data[index] += value;
+ }
+
+ @Override
+ public double get(int index) {
+ check(index);
+ return data[index];
+ }
+
+ @Override
+ public DenseVector copy() {
+ return new DenseVector(this);
+ }
+
+ @Override
+ public DenseVector zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ @Override
+ public DenseVector scale(double alpha) {
+ for (int i = 0; i < size; ++i)
+ data[i] *= alpha;
+ return this;
+ }
+
+ @Override
+ public Vector set(Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.set(y);
+
+ checkSize(y);
+
+ double[] yd = ((DenseVector) y).getData();
+ System.arraycopy(yd, 0, data, 0, size);
+
+ return this;
+ }
+
+ @Override
+ public Vector set(double alpha, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.set(alpha, y);
+
+ checkSize(y);
+
+ if (alpha == 0)
+ return zero();
+
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < size; ++i)
+ data[i] = alpha * yd[i];
+
+ return this;
+ }
+
+ @Override
+ public Vector add(Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.add(y);
+
+ checkSize(y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < size; i++)
+ data[i] += yd[i];
+
+ return this;
+ }
+
+ @Override
+ public Vector add(double alpha, Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.add(alpha, y);
+
+ checkSize(y);
+
+ if (alpha == 0)
+ return this;
+
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < size; i++)
+ data[i] += alpha * yd[i];
+
+ return this;
+ }
+
+ @Override
+ public double dot(Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.dot(y);
+
+ checkSize(y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ double dot = 0.;
+ for (int i = 0; i < size; ++i)
+ dot += data[i] * yd[i];
+ return dot;
+ }
+
+ @Override
+ protected double norm1() {
+ double sum = 0;
+ for (int i = 0; i < size; ++i)
+ sum += Math.abs(data[i]);
+ return sum;
+ }
+
+ @Override
+ protected double norm2() {
+ double norm = 0;
+ for (int i = 0; i < size; ++i)
+ norm += data[i] * data[i];
+ return Math.sqrt(norm);
+ }
+
+ @Override
+ protected double norm2_robust() {
+ double scale = 0, ssq = 1;
+ for (int i = 0; i < size; ++i)
+ if (data[i] != 0) {
+ double absxi = Math.abs(data[i]);
+ if (scale < absxi) {
+ ssq = 1 + ssq * (scale / absxi) * (scale / absxi);
+ scale = absxi;
+ } else
+ ssq += (absxi / scale) * (absxi / scale);
+ }
+ return scale * Math.sqrt(ssq);
+ }
+
+ @Override
+ protected double normInf() {
+ double max = 0;
+ for (int i = 0; i < size; ++i)
+ max = Math.max(Math.abs(data[i]), max);
+ return max;
+ }
+
+ /**
+ * Returns the internal vector contents. The array indices correspond to the
+ * vector indices
+ */
+ public double[] getData() {
+ return data;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/DenseVectorSub.java b/src/main/java/no/uib/cipr/matrix/DenseVectorSub.java
new file mode 100644
index 0000000..40eace8
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/DenseVectorSub.java
@@ -0,0 +1,45 @@
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+
+/**
+ * Wraps a DenseVector, allowing easy access to a sub array of
+ * the original without taking copies.
+ * <p/>
+ * It should be possible to utilise BLAS / LAPACK in various
+ * matrix classes. However, as it would be a mammoth task, it
+ * will be done on an as-needed basis.
+ *
+ * @author Sam Halliday
+ */
+public class DenseVectorSub extends AbstractVector {
+
+ private DenseVector wrapped;
+ private int offset;
+
+ public DenseVectorSub(DenseVector wrapped, int offset, int size) {
+ super(size);
+ if (offset + size > wrapped.size)
+ throw new IllegalArgumentException(offset + "+" + size + ">" + wrapped.size);
+ this.offset = offset;
+ this.wrapped = wrapped;
+ }
+
+ @Override
+ public double get(int index) {
+ check(index);
+ return wrapped.get(offset + index);
+ }
+
+ @Override
+ public void set(int index, double value) {
+ check(index);
+ wrapped.set(offset + index, value);
+ }
+
+ @Override
+ public DenseVector copy() {
+ double[] data = Arrays.copyOfRange(wrapped.getData(), offset, offset + size);
+ return new DenseVector(data, false);
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Diag.java b/src/main/java/no/uib/cipr/matrix/Diag.java
new file mode 100644
index 0000000..7f45d3b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Diag.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/** Diagonal enumeration */
+enum Diag {
+ /** Matrix is not unit diagonal */
+ NonUnit,
+
+ /** Matrix is unit diagonal */
+ Unit;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ if (this == NonUnit)
+ return "N";
+ return "U";
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/EVD.java b/src/main/java/no/uib/cipr/matrix/EVD.java
new file mode 100644
index 0000000..0166dd1
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/EVD.java
@@ -0,0 +1,201 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes eigenvalue decompositions of general matrices
+ */
+public class EVD {
+
+ /**
+ * Double work array
+ */
+ private final double[] work;
+
+ /**
+ * Size of the matrix
+ */
+ private final int n;
+
+ /**
+ * Job to do on the left and right eigenvectors
+ */
+ private final JobEig jobLeft, jobRight;
+
+ /**
+ * Contains the real and imaginary parts of the eigenvalues
+ */
+ private final double[] Wr, Wi;
+
+ /**
+ * Contains the left and the right eigenvectors
+ */
+ private final DenseMatrix Vl, Vr;
+
+ /**
+ * Creates an empty eigenvalue decomposition which will compute all the
+ * eigenvalues and eigenvectors (left and right)
+ *
+ * @param n
+ * Size of the matrix
+ */
+ public EVD(int n) {
+ this(n, true, true);
+ }
+
+ /**
+ * Creates an empty eigenvalue decomposition
+ *
+ * @param n
+ * Size of the matrix
+ * @param left
+ * Whether to compute the left eigenvectors or not
+ * @param right
+ * Whether to compute the right eigenvectors or not
+ */
+ public EVD(int n, boolean left, boolean right) {
+ this.n = n;
+ this.jobLeft = left ? JobEig.All : JobEig.Eigenvalues;
+ this.jobRight = right ? JobEig.All : JobEig.Eigenvalues;
+
+ // Allocate space for the decomposition
+ Wr = new double[n];
+ Wi = new double[n];
+
+ if (left)
+ Vl = new DenseMatrix(n, n);
+ else
+ Vl = null;
+
+ if (right)
+ Vr = new DenseMatrix(n, n);
+ else
+ Vr = null;
+
+ // Find the needed workspace
+ double[] worksize = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeev(jobLeft.netlib(), jobRight.netlib(), n, new double[0],
+ Matrices.ld(n), new double[0], new double[0], new double[0], Matrices.ld(n),
+ new double[0], Matrices.ld(n), worksize, -1, info);
+
+ // Allocate workspace
+ int lwork = 0;
+ if (info.val != 0) {
+ if (jobLeft == JobEig.All || jobRight == JobEig.All)
+ lwork = 4 * n;
+ else
+ lwork = 3 * n;
+ } else
+ lwork = (int) worksize[0];
+
+ lwork = Math.max(1, lwork);
+ work = new double[lwork];
+ }
+
+ /**
+ * Convenience method for computing the complete eigenvalue decomposition of
+ * the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Not modified
+ * @return Newly allocated decomposition
+ * @throws NotConvergedException
+ */
+ public static EVD factorize(Matrix A) throws NotConvergedException {
+ return new EVD(A.numRows()).factor(new DenseMatrix(A));
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current decomposition
+ * @throws NotConvergedException
+ */
+ public EVD factor(DenseMatrix A) throws NotConvergedException {
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+ else if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeev(jobLeft.netlib(), jobRight.netlib(), n, A.getData(),
+ Matrices.ld(n), Wr, Wi, jobLeft == JobEig.All ? Vl.getData() : new double[0],
+ Matrices.ld(n), jobRight == JobEig.All ? Vr.getData() : new double[0], Matrices.ld(n),
+ work, work.length, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+ /**
+ * Gets the left eigenvectors, if available
+ */
+ public DenseMatrix getLeftEigenvectors() {
+ return Vl;
+ }
+
+ /**
+ * Gets the right eigenvectors, if available
+ */
+ public DenseMatrix getRightEigenvectors() {
+ return Vr;
+ }
+
+ /**
+ * Gets the real part of the eigenvalues
+ */
+ public double[] getRealEigenvalues() {
+ return Wr;
+ }
+
+ /**
+ * Gets the imaginary part of the eigenvalues
+ */
+ public double[] getImaginaryEigenvalues() {
+ return Wi;
+ }
+
+ /**
+ * True if the left eigenvectors have been computed
+ */
+ public boolean hasLeftEigenvectors() {
+ return Vl != null;
+ }
+
+ /**
+ * True if the right eigenvectors have been computed
+ */
+ public boolean hasRightEigenvectors() {
+ return Vr != null;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/GivensRotation.java b/src/main/java/no/uib/cipr/matrix/GivensRotation.java
new file mode 100644
index 0000000..18f0bd6
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/GivensRotation.java
@@ -0,0 +1,94 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Givens plane rotation
+ */
+public class GivensRotation {
+
+ /**
+ * Cosine and sine of the rotation angle. c = x / sqrt(x^2 + y^2), and s =
+ * -y / sqrt(x^2 + y^2)
+ */
+ private final double c, s;
+
+ /**
+ * Constructs a Givens plane rotation for a given 2-vector
+ *
+ * @param x
+ * First component of the vector
+ * @param y
+ * Second component of the vector
+ */
+ public GivensRotation(double x, double y) {
+ double roe = Math.abs(x) > Math.abs(y) ? x : y;
+
+ double scale = Math.abs(x) + Math.abs(y);
+ if (scale != 0) {
+ double xs = x / scale;
+ double ys = y / scale;
+ double r = scale * Math.sqrt(xs * xs + ys * ys);
+ if (roe < 0)
+ r *= -1;
+ c = x / r;
+ s = y / r;
+ } else {
+ c = 1;
+ s = 0;
+ }
+ }
+
+ /**
+ * Applies the Givens rotation to two elements in a matrix column
+ *
+ * @param H
+ * Matrix to apply to
+ * @param column
+ * Column index
+ * @param i1
+ * Row index of first element
+ * @param i2
+ * Row index of second element
+ */
+ public void apply(Matrix H, int column, int i1, int i2) {
+ double temp = c * H.get(i1, column) + s * H.get(i2, column);
+ H.set(i2, column, -s * H.get(i1, column) + c * H.get(i2, column));
+ H.set(i1, column, temp);
+ }
+
+ /**
+ * Applies the Givens rotation to two elements of a vector
+ *
+ * @param x
+ * Vector to apply to
+ * @param i1
+ * Index of first element
+ * @param i2
+ * Index of second element
+ */
+ public void apply(Vector x, int i1, int i2) {
+ double temp = c * x.get(i1) + s * x.get(i2);
+ x.set(i2, -s * x.get(i1) + c * x.get(i2));
+ x.set(i1, temp);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/JobEig.java b/src/main/java/no/uib/cipr/matrix/JobEig.java
new file mode 100644
index 0000000..fc8e01b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/JobEig.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/** The job the eigenvectors solvers are to do */
+enum JobEig {
+ /** Compute eigenvalues and eigenvectors */
+ All,
+
+ /** Only compute the eigenvalues */
+ Eigenvalues;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ if (this == All)
+ return "V";
+ return "N";
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/JobEigRange.java b/src/main/java/no/uib/cipr/matrix/JobEigRange.java
new file mode 100644
index 0000000..7441e06
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/JobEigRange.java
@@ -0,0 +1,47 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/** When computing eigenvalues, this indicates which eigenvalues to locate. */
+enum JobEigRange {
+ /** All eigenvalues will be computed */
+ All,
+
+ /** The eigenvalues with the given indices are computed */
+ Indices,
+
+ /** Eigenvalues in a given interval will be found */
+ Interval;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ switch (this) {
+ case All:
+ return "A";
+ case Indices:
+ return "I";
+ default:
+ return "V";
+ }
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/JobSVD.java b/src/main/java/no/uib/cipr/matrix/JobSVD.java
new file mode 100644
index 0000000..f3adf8c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/JobSVD.java
@@ -0,0 +1,62 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/**
+ * The job the singular value solvers are to do. This only limits which singular vectors
+ * are computed, all the singular values are always computed
+ */
+enum JobSVD {
+ /** Compute all of the singular vectors */
+ All,
+
+ /** Do not compute any singular vectors */
+ None,
+
+ /**
+ * Overwrite passed data. For an <code>M*N</code> matrix, this either overwrites the
+ * passed matrix with as many singular vectors as there is room for. Details depend on
+ * the actual algorithm
+ */
+ Overwrite,
+
+ /**
+ * Compute parts of the singular vectors. For an <code>M*N</code> matrix, this
+ * computes <code>min(M,N)</code> singular vectors
+ */
+ Part;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ switch (this) {
+ case All:
+ return "A";
+ case Part:
+ return "S";
+ case Overwrite:
+ return "O";
+ default:
+ return "N";
+ }
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/LQ.java b/src/main/java/no/uib/cipr/matrix/LQ.java
new file mode 100644
index 0000000..d24f97c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LQ.java
@@ -0,0 +1,138 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes LQ decompositions
+ */
+public class LQ extends OrthogonalComputer {
+
+ /**
+ * Constructs an empty LQ decomposition
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns. Must be larger than or equal the number of
+ * rows
+ */
+ public LQ(int m, int n) {
+ super(m, n, false);
+
+ if (n < m)
+ throw new IllegalArgumentException("n < m");
+
+ int lwork;
+
+ // Query optimal workspace. First for computing the factorization
+ {
+ work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgelqf(m, n, new double[0], Matrices.ld(m), new double[0],
+ work, -1, info);
+
+ if (info.val != 0)
+ lwork = m;
+ else
+ lwork = (int) work[0];
+ lwork = Math.max(1, lwork);
+ work = new double[lwork];
+ }
+
+ // Workspace needed for generating an explicit orthogonal matrix
+ {
+ workGen = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dorglq(m, n, m, new double[0],
+ Matrices.ld(m), new double[0], workGen, -1, info);
+
+ if (info.val != 0)
+ lwork = m;
+ else
+ lwork = (int) workGen[0];
+ lwork = Math.max(1, lwork);
+ workGen = new double[lwork];
+ }
+
+ }
+
+ /**
+ * Convenience method to compute a LQ decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return Newly allocated decomposition
+ */
+ public static LQ factorize(Matrix A) {
+ return new LQ(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ @Override
+ public LQ factor(DenseMatrix A) {
+
+ if (Q.numRows() != A.numRows())
+ throw new IllegalArgumentException("Q.numRows() != A.numRows()");
+ else if (Q.numColumns() != A.numColumns())
+ throw new IllegalArgumentException(
+ "Q.numColumns() != A.numColumns()");
+ else if (L == null)
+ throw new IllegalArgumentException("L == null");
+
+ /*
+ * Calculate factorisation, and extract the triangular factor
+ */
+ intW info = new intW(0);
+ LAPACK.getInstance().dgelqf(m, n, A.getData(), Matrices.ld(m), tau, work,
+ work.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ L.zero();
+ for (MatrixEntry e : A)
+ if (e.row() >= e.column())
+ L.set(e.row(), e.column(), e.get());
+
+ /*
+ * Generate the orthogonal matrix
+ */
+ info.val = 0;
+ LAPACK.getInstance().dorglq(m, n, k, A.getData(), Matrices.ld(m), tau, workGen,
+ workGen.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ Q.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the lower triangular factor
+ */
+ public LowerTriangDenseMatrix getL() {
+ return L;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSPDBandMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSPDBandMatrix.java
new file mode 100644
index 0000000..e2c1f44
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSPDBandMatrix.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Lower symmetrical positive definite banded matrix. It does not enforce this
+ * property (except for symmetry), and has the same storage layout as
+ * {@link no.uib.cipr.matrix.LowerSymmBandMatrix LowerSymmBandMatrix}.
+ */
+public class LowerSPDBandMatrix extends LowerSymmBandMatrix {
+
+ /**
+ * Constructor for LowerSPDBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public LowerSPDBandMatrix(int n, int kd) {
+ super(n, kd);
+ }
+
+ /**
+ * Constructor for LowerSPDBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public LowerSPDBandMatrix(Matrix A, int kd) {
+ super(A, kd);
+ }
+
+ /**
+ * Constructor for LowerSPDBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public LowerSPDBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, deep);
+ }
+
+ @Override
+ public LowerSPDBandMatrix copy() {
+ return new LowerSPDBandMatrix(this, kd);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSPDDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSPDDenseMatrix.java
new file mode 100644
index 0000000..f277cf4
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSPDDenseMatrix.java
@@ -0,0 +1,77 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Lower symmetrical positive definite dense matrix. Same layout as
+ * {@link no.uib.cipr.matrix.LowerSymmDenseMatrix LowerSymmDenseMatrix}. This
+ * class does not enforce the SPD property, but serves as a tag so that more
+ * efficient algorithms can be used in the solvers.
+ */
+public class LowerSPDDenseMatrix extends LowerSymmDenseMatrix {
+
+ /**
+ * Constructor for LowerSPDDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerSPDDenseMatrix(int n) {
+ super(n);
+ }
+
+ /**
+ * Constructor for LowerSPDDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the lower
+ * triangular part is copied
+ */
+ public LowerSPDDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for LowerSPDDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the lower
+ * triangular part is copied
+ * @param deep
+ * False for a shallow copy, else it'll be a deep copy. For
+ * shallow copies, <code>A</code> must be a dense matrix
+ */
+ public LowerSPDDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ @Override
+ public LowerSPDDenseMatrix copy() {
+ return new LowerSPDDenseMatrix(this);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSPDPackMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSPDPackMatrix.java
new file mode 100644
index 0000000..e94157c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSPDPackMatrix.java
@@ -0,0 +1,77 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Lower symmetrical positive definite packed matrix. Same layout as
+ * {@link no.uib.cipr.matrix.LowerSymmPackMatrix LowerSymmPackMatrix}. This
+ * class does not enforce the SPD property, but serves as a tag so that more
+ * efficient algorithms can be used in the solvers.
+ */
+public class LowerSPDPackMatrix extends LowerSymmPackMatrix {
+
+ /**
+ * Constructor for LowerSPDPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerSPDPackMatrix(int n) {
+ super(n);
+ }
+
+ /**
+ * Constructor for LowerSPDPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public LowerSPDPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for LowerSPDPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public LowerSPDPackMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ @Override
+ public LowerSPDPackMatrix copy() {
+ return new LowerSPDPackMatrix(this);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSymmBandMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSymmBandMatrix.java
new file mode 100644
index 0000000..eaaf190
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSymmBandMatrix.java
@@ -0,0 +1,99 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower symmetrical banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.BandMatrix BandMatrix}, but without
+ * superdiagonals. Upper part of the matrix is implictly known by symmetry
+ */
+public class LowerSymmBandMatrix extends AbstractSymmBandMatrix {
+
+ /**
+ * Constructor for LowerSymmBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public LowerSymmBandMatrix(int n, int kd) {
+ super(n, kd, 0, UpLo.Lower);
+ }
+
+ /**
+ * Constructor for LowerSymmBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public LowerSymmBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for LowerSymmBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public LowerSymmBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, 0, deep, UpLo.Lower);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column <= row)
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column > row)
+ return super.get(column, row);
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column <= row)
+ super.set(row, column, value);
+ }
+
+ @Override
+ public LowerSymmBandMatrix copy() {
+ return new LowerSymmBandMatrix(this, kd);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSymmDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSymmDenseMatrix.java
new file mode 100644
index 0000000..6c68a6a
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSymmDenseMatrix.java
@@ -0,0 +1,99 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower symmetric dense matrix. It has the same storage layout as the
+ * {@link no.uib.cipr.matrix.DenseMatrix DenseMatrix}, but only refers to
+ * elements below or on the main diagonal. The remaining elements are never
+ * accessed nor changed, and is known only by symmetry.
+ */
+public class LowerSymmDenseMatrix extends AbstractSymmDenseMatrix {
+
+ /**
+ * Constructor for LowerSymmDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerSymmDenseMatrix(int n) {
+ super(n, UpLo.Lower);
+ }
+
+ /**
+ * Constructor for LowerSymmDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the lower
+ * triangular part is copied
+ */
+ public LowerSymmDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for LowerSymmDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the lower
+ * triangular part is copied
+ * @param deep
+ * If false, a shallow copy is made. In that case, <code>A</code>
+ * must be a dense matrix
+ */
+ public LowerSymmDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Lower);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column <= row)
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column > row)
+ return super.get(column, row);
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column <= row)
+ super.set(row, column, value);
+ }
+
+ @Override
+ public LowerSymmDenseMatrix copy() {
+ return new LowerSymmDenseMatrix(this);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() >= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerSymmPackMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerSymmPackMatrix.java
new file mode 100644
index 0000000..5cf13cf
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerSymmPackMatrix.java
@@ -0,0 +1,106 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower symmetric packed matrix. Same storage as
+ * {@link no.uib.cipr.matrix.LowerTriangPackMatrix LowerTriangPackMatrix}, but
+ * the upper triangular part is known by symmetry.
+ */
+public class LowerSymmPackMatrix extends AbstractSymmPackMatrix {
+
+ /**
+ * Constructor for LowerSymmPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerSymmPackMatrix(int n) {
+ super(n, UpLo.Lower);
+ }
+
+ /**
+ * Constructor for LowerSymmPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public LowerSymmPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for LowerSymmPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public LowerSymmPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Lower);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column <= row)
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column <= row)
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column <= row)
+ return data[getIndex(row, column)];
+ return data[getIndex(column, row)];
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + (2 * n - (column + 1)) * column / 2;
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() >= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public LowerSymmPackMatrix copy() {
+ return new LowerSymmPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerTriangBandMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerTriangBandMatrix.java
new file mode 100644
index 0000000..c35a1a9
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerTriangBandMatrix.java
@@ -0,0 +1,110 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower triangular banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.BandMatrix BandMatrix}, but without
+ * superdiagonals.
+ */
+public class LowerTriangBandMatrix extends AbstractTriangBandMatrix {
+
+ /**
+ * Constructor for LowerTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public LowerTriangBandMatrix(int n, int kd) {
+ super(n, kd, 0, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public LowerTriangBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for LowerTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public LowerTriangBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, 0, deep, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ LowerTriangBandMatrix(int n, int kd, Diag diag) {
+ super(n, kd, 0, UpLo.Lower, diag);
+ }
+
+ /**
+ * Constructor for LowerTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ LowerTriangBandMatrix(Matrix A, int kd, boolean deep, Diag diag) {
+ super(A, kd, 0, deep, UpLo.Lower, diag);
+ }
+
+ @Override
+ public LowerTriangBandMatrix copy() {
+ return new LowerTriangBandMatrix(this, kl);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerTriangDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerTriangDenseMatrix.java
new file mode 100644
index 0000000..9e73d63
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerTriangDenseMatrix.java
@@ -0,0 +1,179 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower triangular dense matrix. It has the same storage layout as the
+ * {@link no.uib.cipr.matrix.DenseMatrix DenseMatrix}, but only refers to
+ * elements below or on the main diagonal. The remaining elements are assumed to
+ * be zero, but since they are never accessed, they need not be.
+ */
+public class LowerTriangDenseMatrix extends AbstractTriangDenseMatrix {
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerTriangDenseMatrix(int n) {
+ super(n, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ LowerTriangDenseMatrix(int n, Diag diag) {
+ super(n, UpLo.Lower, diag);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ */
+ public LowerTriangDenseMatrix(Matrix A) {
+ this(A, Math.min(A.numRows(), A.numColumns()));
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ public LowerTriangDenseMatrix(Matrix A, boolean deep) {
+ this(A, Math.min(A.numRows(), A.numColumns()), deep);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ LowerTriangDenseMatrix(Matrix A, boolean deep, Diag diag) {
+ this(A, Math.min(A.numRows(), A.numColumns()), deep, diag);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ */
+ public LowerTriangDenseMatrix(Matrix A, int k) {
+ this(A, k, true);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ public LowerTriangDenseMatrix(Matrix A, int k, boolean deep) {
+ super(A, k, deep, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the lower triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ LowerTriangDenseMatrix(Matrix A, int k, boolean deep, Diag diag) {
+ super(A, k, deep, UpLo.Lower, diag);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column > row)
+ throw new IllegalArgumentException("column > row");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column > row)
+ return 0;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column > row)
+ throw new IllegalArgumentException("column > row");
+ super.set(row, column, value);
+ }
+
+ @Override
+ public LowerTriangDenseMatrix copy() {
+ return new LowerTriangDenseMatrix(this);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() >= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public Matrix set(Matrix A) {
+ zero();
+ copy(A);
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/LowerTriangPackMatrix.java b/src/main/java/no/uib/cipr/matrix/LowerTriangPackMatrix.java
new file mode 100644
index 0000000..cffcb3a
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/LowerTriangPackMatrix.java
@@ -0,0 +1,181 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Lower triangular packed matrix. In contrast with
+ * {@link no.uib.cipr.matrix.LowerTriangDenseMatrix LowerTriangDenseMatrix},
+ * this matrix exploits the sparsity by only storing about half the matrix. As
+ * such, the triangular matrix
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td> </td>
+ * <td> </td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td> </td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td> </td>
+ * </tr>
+ * <tr>
+ * <td>a<sub>41</sub></td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ * <p>
+ * is packed as follows:
+ * </p>
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>21</sub></td>
+ * <td>a<sub>31</sub></td>
+ * <td>a<sub>41</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>32</sub></td>
+ * <td>a<sub>42</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>43</sub></td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ */
+public class LowerTriangPackMatrix extends AbstractTriangPackMatrix {
+
+ /**
+ * Constructor for LowerTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public LowerTriangPackMatrix(int n) {
+ super(n, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ LowerTriangPackMatrix(int n, Diag diag) {
+ super(n, UpLo.Lower, diag);
+ }
+
+ /**
+ * Constructor for LowerTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public LowerTriangPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for LowerTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public LowerTriangPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Lower, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for LowerTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ LowerTriangPackMatrix(Matrix A, boolean deep, Diag diag) {
+ super(A, deep, UpLo.Lower, diag);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column > row)
+ throw new IllegalArgumentException("column > row");
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column > row)
+ throw new IllegalArgumentException("column > row");
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column > row)
+ return 0;
+ return data[getIndex(row, column)];
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + (2 * n - (column + 1)) * column / 2;
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() >= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public LowerTriangPackMatrix copy() {
+ return new LowerTriangPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Matrices.java b/src/main/java/no/uib/cipr/matrix/Matrices.java
new file mode 100644
index 0000000..ef8ea34
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Matrices.java
@@ -0,0 +1,690 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+
+/**
+ * Static utility methods for matrices and vectors
+ */
+public final class Matrices {
+
+ private Matrices() {
+ // No need to instantiate
+ }
+
+ /**
+ * <code>max(1, M)</code> provided as a convenience for 'leading dimension' calculations.
+ *
+ * @param n
+ */
+ static int ld(int n) {
+ return Math.max(1, n);
+ }
+
+ /**
+ * <code>max(1, max(M, N))</code> provided as a convenience for 'leading dimension'
+ * calculations.
+ *
+ * @param m
+ * @param n
+ */
+ static int ld(int m, int n) {
+ return Math.max(1, Math.max(m, n));
+ }
+
+ /**
+ * Returns the number of non-zero entries in the given vector
+ */
+ public static int cardinality(Vector x) {
+ int nz = 0;
+ for (VectorEntry e : x)
+ if (e.get() != 0)
+ nz++;
+ return nz;
+ }
+
+ /**
+ * Returns the number of non-zero entries in the given matrix
+ */
+ public static int cardinality(Matrix A) {
+ int nz = 0;
+ for (MatrixEntry e : A)
+ if (e.get() != 0)
+ nz++;
+ return nz;
+ }
+
+ /**
+ * Returns an array of arrays containing a copy of the given matrix. Each
+ * array contains one row.
+ */
+ public static double[][] getArray(Matrix A) {
+ double[][] Ad = new double[A.numRows()][A.numColumns()];
+ for (MatrixEntry e : A)
+ Ad[e.row()][e.column()] = e.get();
+ return Ad;
+ }
+
+ /**
+ * Returns a dense array containing a copy of the given vector
+ */
+ public static double[] getArray(Vector x) {
+ double[] xd = new double[x.size()];
+ for (VectorEntry e : x)
+ xd[e.index()] = e.get();
+ return xd;
+ }
+
+ /**
+ * Returns the identity matrix of the given size
+ *
+ * @param size
+ * Number of rows/columns of the matrix
+ * @return Matrix of the given size, with ones on the main diagonal
+ */
+ public static DenseMatrix identity(int size) {
+ DenseMatrix A = new DenseMatrix(size, size);
+ for (int i = 0; i < size; ++i)
+ A.set(i, i, 1);
+ return A;
+ }
+
+ /**
+ * Creates a random vector. Numbers are drawn from a uniform distribution
+ * between 0 and 1
+ *
+ * @param size
+ * Size of the vector
+ */
+ public static Vector random(int size) {
+ return random(new DenseVector(size));
+ }
+
+ /**
+ * Populates a vector with random numbers drawn from a uniform distribution
+ * between 0 and 1
+ *
+ * @param x
+ * Vector to populate
+ */
+ public static Vector random(Vector x) {
+ for (int i = 0; i < x.size(); ++i)
+ x.set(i, Math.random());
+ return x;
+ }
+
+ /**
+ * Creates a random matrix. Numbers are drawn from a uniform distribution
+ * between 0 and 1
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of columns
+ */
+ public static Matrix random(int numRows, int numColumns) {
+ return random(new DenseMatrix(numRows, numColumns));
+ }
+
+ /**
+ * Populates a matrix with random numbers drawn from a uniform distribution
+ * between 0 and 1
+ *
+ * @param A
+ * Matrix to populate
+ */
+ public static Matrix random(Matrix A) {
+ for (int j = 0; j < A.numColumns(); ++j)
+ for (int i = 0; i < A.numRows(); ++i)
+ A.set(i, j, Math.random());
+ return A;
+ }
+
+ /**
+ * Returns a synchronized vector which wraps the given vector. Only the
+ * <code>set(int, double)</code> and <code>add(int, double)</code>
+ * methods and their blocked versions are synchronized.
+ * <p>
+ * <b>Note: </b> Do not use the wrapped vector for any operations besides
+ * matrix assembly, as these operations may be very slow.
+ *
+ * @param x
+ * Vector to be wrapped
+ * @return A thin wrapper around <code>x</code>
+ */
+ public static Vector synchronizedVector(Vector x) {
+ return new SynchronizedVector(x);
+ }
+
+ /**
+ * Returns a synchronized matrix which wraps the given matrix. Only the
+ * <code>set(int, int, double)</code> and
+ * <code>add(int, int, double)</code> methods and their blocked versions
+ * are synchronized.
+ * <p>
+ * <b>Note: </b> Do not use the wrapped matrix for any operations besides
+ * matrix assembly, as these operations may be very slow.
+ *
+ * @param A
+ * Matrix to be wrapped
+ * @return A thin wrapper around <code>A</code>
+ */
+ public static Matrix synchronizedMatrix(Matrix A) {
+ return new SynchronizedMatrix(A);
+ }
+
+ /**
+ * Returns a synchronized matrix which wraps the given matrix. Only the
+ * <code>set(int, int, double)</code> and
+ * <code>add(int, int, double)</code> methods and their blocked versions
+ * are synchronized.
+ * <p>
+ * The locking provided is finer than the locking of the whole matrix, as
+ * different threads can access different rows simultaneous, while only one
+ * thread can access a given row at a time. Use this for row-major matrices,
+ * <i>not </i> for column-major matrices.
+ * <p>
+ * <b>Note: </b> Do not use the wrapped matrix for any operations besides
+ * matrix assembly, as these operations may be very slow.
+ *
+ * @param A
+ * Matrix to be wrapped
+ * @return A thin wrapper around <code>A</code>. Individual rows are
+ * locked
+ */
+ public static Matrix synchronizedMatrixByRows(Matrix A) {
+ return new SynchronizedRowMatrix(A);
+ }
+
+ /**
+ * Returns a synchronized matrix which wraps the given matrix. Only the
+ * <code>set(int, int, double)</code> and
+ * <code>add(int, int, double)</code> methods and their blocked versions
+ * are synchronized.
+ * <p>
+ * The locking provided is finer than the locking of the whole matrix, as
+ * different threads can access different columns simultaneous, while only
+ * one thread can access a given column at a time. Use this for column-major
+ * matrices, <i>not </i> for row-major matrices.
+ * <p>
+ * <b>Note: </b> Do not use the wrapped matrix for any operations besides
+ * matrix assembly, as these operations may be very slow.
+ *
+ * @param A
+ * Matrix to be wrapped
+ * @return A thin wrapper around <code>A</code>. Individual columns are
+ * locked
+ */
+ public static Matrix synchronizedMatrixByColumns(Matrix A) {
+ return new SynchronizedColumnMatrix(A);
+ }
+
+ /**
+ * Returns a view into the given matrix. This view is only for easing some
+ * matrix-assembly cases, not for general use. To extract a more
+ * higher-performing and general matrix, create a copy of the submatrix. The
+ * result is a {@link no.uib.cipr.matrix.DenseMatrix DenseMatrix}.
+ *
+ * @param A
+ * Matrix to create view on
+ * @param row
+ * Rows to access. Must be within the bounds of <code>A</code>
+ * @param column
+ * Columns to access. Must be within the bounds of <code>A</code>
+ * @return Submatrix of <code>A</code>. Changing it will change the
+ * backing matrix
+ */
+ public static Matrix getSubMatrix(Matrix A, int[] row, int[] column) {
+ return new RefMatrix(A, row, column);
+ }
+
+ /**
+ * Returns a view into the given vector. This view is only for easing some
+ * vector-assembly cases, not for general use. To extract a more
+ * higher-performing and general vector, create a copy of the subvector. The
+ * result is a {@link no.uib.cipr.matrix.DenseVector DenseVector}.
+ *
+ * @param x
+ * Vector to create view on
+ * @param index
+ * Indices to access. Must be within the bounds of <code>x</code>
+ * @return Submatrix of <code>x</code>. Changing it will change the
+ * backing matrix
+ */
+ public static Vector getSubVector(Vector x, int[] index) {
+ return new RefVector(x, index);
+ }
+
+ /**
+ * Matrix backed by another matrix. Used by <code>getSubMatrix</code>
+ */
+ private static class RefMatrix extends AbstractMatrix {
+
+ private Matrix A;
+
+ private int[] row, column;
+
+ public RefMatrix(Matrix A, int[] row, int[] column) {
+ super(row.length, column.length);
+ this.A = A;
+ this.row = row;
+ this.column = column;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ A.add(this.row[row], this.column[column], value);
+ }
+
+ @Override
+ public DenseMatrix copy() {
+ return new DenseMatrix(this);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return A.get(this.row[row], this.column[column]);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ A.set(this.row[row], this.column[column], value);
+ }
+
+ }
+
+ /**
+ * Vector backed by another vector. Used by <code>getSubVector</code>
+ */
+ private static class RefVector extends AbstractVector {
+
+ private Vector x;
+
+ private int[] index;
+
+ public RefVector(Vector x, int[] index) {
+ super(index.length);
+ this.x = x;
+ this.index = index;
+ }
+
+ @Override
+ public void add(int index, double value) {
+ x.add(this.index[index], value);
+ }
+
+ @Override
+ public DenseVector copy() {
+ return new DenseVector(this);
+ }
+
+ @Override
+ public double get(int index) {
+ return x.get(this.index[index]);
+ }
+
+ @Override
+ public void set(int index, double value) {
+ x.set(this.index[index], value);
+ }
+
+ }
+
+ /**
+ * Ensures correctness in the vector assembly. Since it extends the
+ * AbstractVector class, algebraic operations will be slow. It is not
+ * possible to implement Vector and delegate calls to the imbedded vector,
+ * since casting to the imbedded vector is not possible
+ */
+ private static class SynchronizedVector extends AbstractVector {
+
+ private Vector x;
+
+ public SynchronizedVector(Vector x) {
+ super(x);
+ this.x = x;
+ }
+
+ @Override
+ public synchronized void add(int index, double value) {
+ x.add(index, value);
+ }
+
+ @Override
+ public synchronized void set(int index, double value) {
+ x.set(index, value);
+ }
+
+ @Override
+ public synchronized double get(int index) {
+ return x.get(index);
+ }
+
+ @Override
+ public Vector copy() {
+ return Matrices.synchronizedVector(x.copy());
+ }
+
+ }
+
+ /**
+ * Ensures correctness in the matrix assembly. Since it extends the
+ * AbstractMatrix class, algebraic operations will be slow. It is not
+ * possible to implement Matrix and delegate calls to the imbedded matrix,
+ * since casting to the imbedded matrix is not possible
+ */
+ private static class SynchronizedMatrix extends AbstractMatrix {
+
+ private Matrix A;
+
+ public SynchronizedMatrix(Matrix A) {
+ super(A);
+ this.A = A;
+ }
+
+ @Override
+ public synchronized void add(int row, int column, double value) {
+ A.add(row, column, value);
+ }
+
+ @Override
+ public synchronized void set(int row, int column, double value) {
+ A.set(row, column, value);
+ }
+
+ @Override
+ public synchronized double get(int row, int column) {
+ return A.get(row, column);
+ }
+
+ @Override
+ public Matrix copy() {
+ return Matrices.synchronizedMatrix(A.copy());
+ }
+
+ }
+
+ /**
+ * Ensures correctness in the matrix assembly. Since it extends the
+ * AbstractMatrix class, algebraic operations will be slow. It is not
+ * possible to implement Matrix and delegate calls to the imbedded matrix,
+ * since casting to the imbedded matrix is not possible
+ * <p>
+ * Locks individual rows instead of the whole matrix
+ */
+ private static class SynchronizedRowMatrix extends AbstractMatrix {
+
+ private Matrix A;
+
+ private Object[] lock;
+
+ public SynchronizedRowMatrix(Matrix A) {
+ super(A);
+ this.A = A;
+ lock = new Object[A.numRows()];
+ for (int i = 0; i < lock.length; ++i)
+ lock[i] = new Object();
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ synchronized (lock[row]) {
+ A.add(row, column, value);
+ }
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ synchronized (lock[row]) {
+ A.set(row, column, value);
+ }
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return A.get(row, column);
+ }
+
+ @Override
+ public Matrix copy() {
+ return Matrices.synchronizedMatrixByRows(A.copy());
+ }
+
+ }
+
+ /**
+ * Ensures correctness in the matrix assembly. Implements matrix instead of
+ * subclassing the abstract matrix in order to correctly delegate every
+ * method to possbly overridden method in the encapsulated matrix.
+ * <p>
+ * Locks individual columns instead of the whole matrix
+ */
+ private static class SynchronizedColumnMatrix extends AbstractMatrix {
+
+ private Matrix A;
+
+ private Object[] lock;
+
+ public SynchronizedColumnMatrix(Matrix A) {
+ super(A);
+ this.A = A;
+ lock = new Object[A.numColumns()];
+ for (int i = 0; i < lock.length; ++i)
+ lock[i] = new Object();
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ synchronized (lock[column]) {
+ A.add(row, column, value);
+ }
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ synchronized (lock[column]) {
+ A.set(row, column, value);
+ }
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return A.get(row, column);
+ }
+
+ @Override
+ public Matrix copy() {
+ return Matrices.synchronizedMatrixByColumns(A.copy());
+ }
+
+ }
+
+ /**
+ * Creates a continuous linear index.
+ *
+ * @param from
+ * Start, inclusive
+ * @param to
+ * Stop, exclusive
+ */
+ public static int[] index(int from, int to) {
+ int length = to - from;
+
+ if (length < 0)
+ length = 0;
+
+ int[] index = new int[length];
+ for (int i = from, j = 0; j < length; ++i, ++j)
+ index[j] = i;
+ return index;
+ }
+
+ /**
+ * Creates a strided linear index.
+ *
+ * @param from
+ * Start, inclusive
+ * @param stride
+ * <code>stride=1</code> for continuous. Negative strides are
+ * allowed
+ * @param to
+ * Stop, exclusive
+ */
+ public static int[] index(int from, int stride, int to) {
+ if (stride == 1)
+ return index(from, to);
+ else if (stride == 0)
+ return new int[0];
+
+ if (to <= from && stride > 0)
+ return new int[0];
+ if (from <= to && stride < 0)
+ return new int[0];
+
+ int length = Math.abs((to - from) / stride);
+ if (Math.abs((to - from) % stride) > 0)
+ length++;
+
+ if (length < 0)
+ length = 0;
+
+ int[] index = new int[length];
+ for (int i = from, j = 0; j < length; i += stride, ++j)
+ index[j] = i;
+ return index;
+ }
+
+ /**
+ * Finds the number of non-zero entries on each row
+ */
+ public static int[] rowBandwidth(Matrix A) {
+ int[] nz = new int[A.numRows()];
+
+ for (MatrixEntry e : A)
+ nz[e.row()]++;
+
+ return nz;
+ }
+
+ /**
+ * Finds the number of non-zero entries on each column
+ */
+ public static int[] columnBandwidth(Matrix A) {
+ int[] nz = new int[A.numColumns()];
+
+ for (MatrixEntry e : A)
+ nz[e.column()]++;
+
+ return nz;
+ }
+
+ /**
+ * Finds the number of diagonals below the main diagonal. Useful for
+ * converting a general matrix into a banded matrix
+ */
+ public static int getNumSubDiagonals(Matrix A) {
+ int kl = 0;
+
+ for (MatrixEntry e : A)
+ kl = Math.max(kl, e.row() - e.column());
+
+ return kl;
+ }
+
+ /**
+ * Finds the number of diagonals above the main diagonal. Useful for
+ * converting a general matrix into a banded matrix
+ */
+ public static int getNumSuperDiagonals(Matrix A) {
+ int ku = 0;
+
+ for (MatrixEntry e : A)
+ ku = Math.max(ku, e.column() - e.row());
+
+ return ku;
+ }
+
+ /**
+ * Sets the selected rows of <code>A</code> equal zero, and puts
+ * <code>diagonal</code> on the diagonal of those rows. Useful for
+ * enforcing boundary conditions
+ */
+ public static void zeroRows(Matrix A, double diagonal, int... row) {
+ // Sort the rows
+ int[] rowS = row.clone();
+ Arrays.sort(rowS);
+
+ for (MatrixEntry e : A) {
+ int j = java.util.Arrays.binarySearch(rowS, e.row());
+ if (j >= 0) { // Found
+ if (e.row() == e.column()) // Diagonal
+ e.set(diagonal);
+ else
+ // Off diagonal
+ e.set(0);
+ }
+ }
+
+ // Ensure the diagonal is set. This is necessary in case of missing
+ // rows
+ if (diagonal != 0)
+ for (int rowI : row)
+ A.set(rowI, rowI, diagonal);
+ }
+
+ /**
+ * Sets the selected columns of <code>A</code> equal zero, and puts
+ * <code>diagonal</code> on the diagonal of those columns. Useful for
+ * enforcing boundary conditions
+ */
+ public static void zeroColumns(Matrix A, double diagonal, int... column) {
+ // Sort the columns
+ int[] columnS = column.clone();
+ Arrays.sort(columnS);
+
+ for (MatrixEntry e : A) {
+ int j = java.util.Arrays.binarySearch(columnS, e.column());
+ if (j >= 0) { // Found
+ if (e.row() == e.column()) // Diagonal
+ e.set(diagonal);
+ else
+ // Off diagonal
+ e.set(0);
+ }
+ }
+
+ // Ensure the diagonal is set. This is necessary in case of missing
+ // columns
+ if (diagonal != 0)
+ for (int columnI : column)
+ A.set(columnI, columnI, diagonal);
+ }
+
+ public static DenseVector getColumn(Matrix m, int j) {
+ DenseVector v = new DenseVector(m.numRows());
+ for (int i = 0; i < v.size(); i++) {
+ v.set(i, m.get(i, j));
+ }
+ return v;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Matrix.java b/src/main/java/no/uib/cipr/matrix/Matrix.java
new file mode 100644
index 0000000..f263a84
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Matrix.java
@@ -0,0 +1,755 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Basic matrix interface. It holds <code>double</code>s in a rectangular 2D
+ * array, and it is used alongside <code>Vector</code> in numerical
+ * computations. Implementing classes decides on the actual storage.
+ *
+ * <h4>Basic operations</h4>
+ * <p>
+ * Use <code>numRows</code> and <code>numColumns</code> to get the basic
+ * size of a matrix. <code>get(int,int)</code> gets an element, and there are
+ * corresponding <code>set(int,int,double)</code> and
+ * <code>add(int,int,double)</code> methods as well. Note that matrix indices
+ * are zero-based (typical for Java and C). This means that the row-indices
+ * range from 0 to <code>numRows-1</code>, likewise for the columns. It is
+ * legal to have <code>numRows</code> or <code>numColumns</code> equal zero.
+ * </p>
+ * <p>
+ * Other basic operations are <code>zero</code> which zeros all the entries of
+ * the matrix, which can be cheaper than either zeroing the matrix manually, or
+ * creating a new matrix, and the operation <code>copy</code> which creates a
+ * deep copy of the matrix. This copy has separate storage, but starts with the
+ * same contents as the current matrix.
+ * </p>
+ *
+ * <h4>Iterators</h4>
+ * <p>
+ * The matrix interface extends <code>Iterable</code>, and the iterator
+ * returns a <code>MatrixEntry</code> which contains current index and entry
+ * value. Note that the iterator may skip non-zero entries. Using an iterator,
+ * many simple and efficient algorithms can be created. The iterator also
+ * permits changing values in the matrix, however only non-zero entries can be
+ * changed.
+ * </p>
+ *
+ * <h4>Basic linear algebra</h4>
+ * <p>
+ * A large selection of basic linear algebra operations are available. To ensure
+ * high efficiency, little or no internal memory allocation is done, and the
+ * user is required to supply the output arguments.
+ * </p>
+ * <p>
+ * The operations available include:
+ * </p>
+ * <dl>
+ * <dt><i>Additions </i></dt>
+ * <dd>Matrices can be added to each other, even if their underlying matrix
+ * structures are different.</dd>
+ * <dt><i>Multiplications </i></dt>
+ * <dd>A matrix can be multiplied with vectors and other matrices. For
+ * increased efficiency, a multiplication can be combined with addition and
+ * scaling, and transpose matrix multiplications are also available.</dd>
+ * <dt><i>Rank-updates </i></dt>
+ * <dd>A matrix can be efficiently updated using low-rank updates. The updates
+ * can be contained in both matrices or vectors.</dd>
+ * <dt><i>Transpositions </i></dt>
+ * <dd>In-place transpositions of square matrices is supported, and the
+ * transpose of a matrix can be stored in another matrix of compatible size
+ * (possibly non-rectangular)</dd>
+ * <dt><i>Solvers </i></dt>
+ * <dd>Many dense and structured sparse matrices have fast, direct solvers, and
+ * can be used to solve linear systems without creating a factorization. These
+ * solvers are typically backed by subroutines in LAPACK</dd>
+ * </dl>
+ */
+public interface Matrix extends Iterable<MatrixEntry> {
+
+ /**
+ * Number of rows in the matrix
+ */
+ int numRows();
+
+ /**
+ * Number of columns in the matrix
+ */
+ int numColumns();
+
+ /**
+ * Returns true if the matrix is square
+ */
+ boolean isSquare();
+
+ /**
+ * <code>A(row,column) = value</code>
+ */
+ void set(int row, int column, double value);
+
+ /**
+ * <code>A(row,column) += value</code>
+ */
+ void add(int row, int column, double value);
+
+ /**
+ * Returns <code>A(row,column)</code>
+ */
+ double get(int row, int column);
+
+ /**
+ * Creates a deep copy of the matrix
+ *
+ * @return A
+ */
+ Matrix copy();
+
+ /**
+ * Zeros all the entries in the matrix, while preserving any underlying
+ * structure. Useful for general, unstructured matrices.
+ *
+ * @return A
+ */
+ Matrix zero();
+
+ /**
+ * <code>y = A*x</code>
+ *
+ * @param x
+ * Vector of size <code>A.numColumns()</code>
+ * @param y
+ * Vector of size <code>A.numRows()</code>
+ * @return y
+ */
+ Vector mult(Vector x, Vector y);
+
+ /**
+ * <code>y = alpha*A*x</code>
+ *
+ * @param x
+ * Vector of size <code>A.numColumns()</code>
+ * @param y
+ * Vector of size <code>A.numRows()</code>
+ * @return y
+ */
+ Vector mult(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>y = A*x + y</code>
+ *
+ * @param x
+ * Vector of size <code>A.numColumns()</code>
+ * @param y
+ * Vector of size <code>A.numRows()</code>
+ * @return y
+ */
+ Vector multAdd(Vector x, Vector y);
+
+ /**
+ * <code>y = alpha*A*x + y</code>
+ *
+ * @param x
+ * Vector of size <code>A.numColumns()</code>
+ * @param y
+ * Vector of size <code>A.numRows()</code>
+ * @return y
+ */
+ Vector multAdd(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>y = A<sup>T</sup>*x</code>
+ *
+ * @param x
+ * Vector of size <code>A.numRows()</code>
+ * @param y
+ * Vector of size <code>A.numColumns()</code>
+ * @return y
+ */
+ Vector transMult(Vector x, Vector y);
+
+ /**
+ * <code>y = alpha*A<sup>T</sup>*x</code>
+ *
+ * @param x
+ * Vector of size <code>A.numRows()</code>
+ * @param y
+ * Vector of size <code>A.numColumns()</code>
+ * @return y
+ */
+ Vector transMult(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>y = A<sup>T</sup>*x + y</code>
+ *
+ * @param x
+ * Vector of size <code>A.numRows()</code>
+ * @param y
+ * Vector of size <code>A.numColumns()</code>
+ * @return y
+ */
+ Vector transMultAdd(Vector x, Vector y);
+
+ /**
+ * <code>y = alpha*A<sup>T</sup>*x + y</code>
+ *
+ * @param x
+ * Vector of size <code>A.numRows()</code>
+ * @param y
+ * Vector of size <code>A.numColumns()</code>
+ * @return y
+ */
+ Vector transMultAdd(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>x = A\b</code>. Not all matrices support this operation, those
+ * that do not throw <code>UnsupportedOperationException</code>. Note
+ * that it is often more efficient to use a matrix decomposition and its
+ * associated solver
+ *
+ * @param b
+ * Vector of size <code>A.numRows()</code>
+ * @param x
+ * Vector of size <code>A.numColumns()</code>
+ * @return x
+ * @throws MatrixSingularException
+ * If the matrix is singular
+ * @throws MatrixNotSPDException
+ * If the solver assumes that the matrix is symmetrical,
+ * positive definite, but that that property does not hold
+ */
+ Vector solve(Vector b, Vector x) throws MatrixSingularException,
+ MatrixNotSPDException;
+
+ /**
+ * <code>x = A<sup>T</sup>\b</code>. Not all matrices support this
+ * operation, those that do not throw
+ * <code>UnsupportedOperationException</code>. Note that it is often more
+ * efficient to use a matrix decomposition and its associated solver
+ *
+ * @param b
+ * Vector of size <code>A.numColumns()</code>
+ * @param x
+ * Vector of size <code>A.numRows()</code>
+ * @return x
+ * @throws MatrixSingularException
+ * If the matrix is singular
+ * @throws MatrixNotSPDException
+ * If the solver assumes that the matrix is symmetrical,
+ * positive definite, but that that property does not hold
+ */
+ Vector transSolve(Vector b, Vector x) throws MatrixSingularException,
+ MatrixNotSPDException;
+
+ /**
+ * <code>A = x*x<sup>T</sup> + A</code>. The matrix must be square, and
+ * the vector of the same length
+ *
+ * @return A
+ */
+ Matrix rank1(Vector x);
+
+ /**
+ * <code>A = alpha*x*x<sup>T</sup> + A</code>. The matrix must be
+ * square, and the vector of the same length
+ *
+ * @return A
+ */
+ Matrix rank1(double alpha, Vector x);
+
+ /**
+ * <code>A = x*y<sup>T</sup> + A</code>. The matrix must be square, and
+ * the vectors of the same length
+ *
+ * @return A
+ */
+ Matrix rank1(Vector x, Vector y);
+
+ /**
+ * <code>A = alpha*x*y<sup>T</sup> + A</code>. The matrix must be
+ * square, and the vectors of the same length
+ *
+ * @return A
+ */
+ Matrix rank1(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>A = x*y<sup>T</sup> + y*x<sup>T</sup> + A</code>. The matrix
+ * must be square, and the vectors of the same length
+ *
+ * @return A
+ */
+ Matrix rank2(Vector x, Vector y);
+
+ /**
+ * <code>A = alpha*x*y<sup>T</sup> + alpha*y*x<sup>T</sup> + A</code>.
+ * The matrix must be square, and the vectors of the same length
+ *
+ * @return A
+ */
+ Matrix rank2(double alpha, Vector x, Vector y);
+
+ /**
+ * <code>C = A*B</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix mult(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A*B</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix mult(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A*B + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix multAdd(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A*B + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix multAdd(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A<sup>T</sup>*B</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transAmult(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A<sup>T</sup>*B</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transAmult(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A<sup>T</sup>*B + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transAmultAdd(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A<sup>T</sup>*B + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transAmultAdd(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A*B<sup>T</sup></code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transBmult(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A*B<sup>T</sup></code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transBmult(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A*B<sup>T</sup> + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transBmultAdd(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A*B<sup>T</sup> + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numRows() == A.numRows()</code> and
+ * <code>B.numColumns() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numColumns() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transBmultAdd(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A<sup>T</sup>*B<sup>T</sup></code>
+ *
+ * @param B
+ * Matrix such that <code>B.numColumns() == A.numRows()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transABmult(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A<sup>T</sup>*B<sup>T</sup></code>
+ *
+ * @param B
+ * Matrix such that <code>B.numColumns() == A.numRows()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transABmult(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>C = A<sup>T</sup>*B<sup>T</sup> + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numColumns() == A.numRows()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transABmultAdd(Matrix B, Matrix C);
+
+ /**
+ * <code>C = alpha*A<sup>T</sup>*B<sup>T</sup> + C</code>
+ *
+ * @param B
+ * Matrix such that <code>B.numColumns() == A.numRows()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @param C
+ * Matrix such that <code>C.numRows() == A.numColumns()</code>
+ * and <code>B.numRows() == C.numColumns()</code>
+ * @return C
+ */
+ Matrix transABmultAdd(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>X = A\B</code>. Not all matrices support this operation, those
+ * that do not throw <code>UnsupportedOperationException</code>. Note
+ * that it is often more efficient to use a matrix decomposition and its
+ * associated solver
+ *
+ * @param B
+ * Matrix with the same number of rows as <code>A</code>, and
+ * the same number of columns as <code>X</code>
+ * @param X
+ * Matrix with a number of rows equal <code>A.numColumns()</code>,
+ * and the same number of columns as <code>B</code>
+ * @return X
+ * @throws MatrixSingularException
+ * If the matrix is singular
+ * @throws MatrixNotSPDException
+ * If the solver assumes that the matrix is symmetrical,
+ * positive definite, but that that property does not hold
+ */
+ Matrix solve(Matrix B, Matrix X) throws MatrixSingularException,
+ MatrixNotSPDException;
+
+ /**
+ * <code>X = A<sup>T</sup>\B</code>. Not all matrices support this
+ * operation, those that do not throw
+ * <code>UnsupportedOperationException</code>. Note that it is often more
+ * efficient to use a matrix decomposition and its associated transpose
+ * solver
+ *
+ * @param B
+ * Matrix with a number of rows equal <code>A.numColumns()</code>,
+ * and the same number of columns as <code>X</code>
+ * @param X
+ * Matrix with the same number of rows as <code>A</code>, and
+ * the same number of columns as <code>B</code>
+ * @return X
+ * @throws MatrixSingularException
+ * If the matrix is singular
+ * @throws MatrixNotSPDException
+ * If the solver assumes that the matrix is symmetrical,
+ * positive definite, but that that property does not hold
+ */
+ Matrix transSolve(Matrix B, Matrix X) throws MatrixSingularException,
+ MatrixNotSPDException;
+
+ /**
+ * <code>A = C*C<sup>T</sup> + A</code>. The matrices must be square
+ * and of the same size
+ *
+ * @return A
+ */
+ Matrix rank1(Matrix C);
+
+ /**
+ * <code>A = alpha*C*C<sup>T</sup> + A</code>. The matrices must be
+ * square and of the same size
+ *
+ * @return A
+ */
+ Matrix rank1(double alpha, Matrix C);
+
+ /**
+ * <code>A = C<sup>T</sup>*C + A</code> The matrices must be square and
+ * of the same size
+ *
+ * @return A
+ */
+ Matrix transRank1(Matrix C);
+
+ /**
+ * <code>A = alpha*C<sup>T</sup>*C + A</code> The matrices must be
+ * square and of the same size
+ *
+ * @return A
+ */
+ Matrix transRank1(double alpha, Matrix C);
+
+ /**
+ * <code>A = B*C<sup>T</sup> + C*B<sup>T</sup> + A</code>. This
+ * matrix must be square
+ *
+ * @param B
+ * Matrix with the same number of rows as <code>A</code> and
+ * the same number of columns as <code>C</code>
+ * @param C
+ * Matrix with the same number of rows as <code>A</code> and
+ * the same number of columns as <code>B</code>
+ * @return A
+ */
+ Matrix rank2(Matrix B, Matrix C);
+
+ /**
+ * <code>A = alpha*B*C<sup>T</sup> + alpha*C*B<sup>T</sup> + A</code>.
+ * This matrix must be square
+ *
+ * @param B
+ * Matrix with the same number of rows as <code>A</code> and
+ * the same number of columns as <code>C</code>
+ * @param C
+ * Matrix with the same number of rows as <code>A</code> and
+ * the same number of columns as <code>B</code>
+ * @return A
+ */
+ Matrix rank2(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>A = B<sup>T</sup>*C + C<sup>T</sup>*B + A</code>. This
+ * matrix must be square
+ *
+ * @param B
+ * Matrix with the same number of rows as <code>C</code> and
+ * the same number of columns as <code>A</code>
+ * @param C
+ * Matrix with the same number of rows as <code>B</code> and
+ * the same number of columns as <code>A</code>
+ * @return A
+ */
+ Matrix transRank2(Matrix B, Matrix C);
+
+ /**
+ * <code>A = alpha*B<sup>T</sup>*C + alpha*C<sup>T</sup>*B + A</code>.
+ * This matrix must be square
+ *
+ * @param B
+ * Matrix with the same number of rows as <code>C</code> and
+ * the same number of columns as <code>A</code>
+ * @param C
+ * Matrix with the same number of rows as <code>B</code> and
+ * the same number of columns as <code>A</code>
+ * @return A
+ */
+ Matrix transRank2(double alpha, Matrix B, Matrix C);
+
+ /**
+ * <code>A = alpha*A</code>
+ *
+ * @return A
+ */
+ Matrix scale(double alpha);
+
+ /**
+ * <code>A=B</code>. The matrices must be of the same size
+ *
+ * @return A
+ */
+ Matrix set(Matrix B);
+
+ /**
+ * <code>A=alpha*B</code>. The matrices must be of the same size
+ *
+ * @return A
+ */
+ Matrix set(double alpha, Matrix B);
+
+ /**
+ * <code>A = B + A</code>. The matrices must be of the same size
+ *
+ * @return A
+ */
+ Matrix add(Matrix B);
+
+ /**
+ * <code>A = alpha*B + A</code>. The matrices must be of the same size
+ *
+ * @return A
+ */
+ Matrix add(double alpha, Matrix B);
+
+ /**
+ * Transposes the matrix in-place. In most cases, the matrix must be square
+ * for this to work.
+ *
+ * @return This matrix
+ */
+ Matrix transpose();
+
+ /**
+ * Sets the tranpose of this matrix into <code>B</code>. Matrix
+ * dimensions must be compatible
+ *
+ * @param B
+ * Matrix with as many rows as this matrix has columns, and as
+ * many columns as this matrix has rows
+ * @return The matrix <code>B=A<sup>T</sup></code>
+ */
+ Matrix transpose(Matrix B);
+
+ /**
+ * Computes the given norm of the matrix
+ *
+ * @param type
+ * The type of norm to compute
+ */
+ double norm(Norm type);
+
+ /**
+ * Supported matrix-norms. Note that <code>Maxvalue</code> is not a proper
+ * matrix norm
+ */
+ enum Norm {
+
+ /**
+ * Maximum absolute row sum
+ */
+ One,
+
+ /**
+ * The root of sum of the sum of squares
+ */
+ Frobenius,
+
+ /**
+ * Maximum column sum
+ */
+ Infinity,
+
+ /**
+ * Largest entry in absolute value. Not a proper matrix norm
+ */
+ Maxvalue;
+
+ /**
+ * @return the String as required by the netlib libraries to represent this norm.
+ */
+ public String netlib() {
+ // TODO: this is a bit of a hack
+ // shouldn't need to know about the internals of netlib
+ if (this == One)
+ return "1";
+ else if (this == Infinity)
+ return "I";
+ else
+ throw new IllegalArgumentException(
+ "Norm must be the 1 or the Infinity norm");
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/MatrixEntry.java b/src/main/java/no/uib/cipr/matrix/MatrixEntry.java
new file mode 100644
index 0000000..82051c3
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/MatrixEntry.java
@@ -0,0 +1,48 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * An entry of a matrix. Returned by the iterators over a matrix structure
+ */
+public interface MatrixEntry {
+
+ /**
+ * Returns the current row index
+ */
+ int row();
+
+ /**
+ * Returns the current column index
+ */
+ int column();
+
+ /**
+ * Returns the value at the current index
+ */
+ double get();
+
+ /**
+ * Sets the value at the current index
+ */
+ void set(double value);
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/MatrixNotSPDException.java b/src/main/java/no/uib/cipr/matrix/MatrixNotSPDException.java
new file mode 100644
index 0000000..02782ac
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/MatrixNotSPDException.java
@@ -0,0 +1,47 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Matrix is not symmetrical, positive definite
+ */
+public class MatrixNotSPDException extends RuntimeException {
+
+ private static final long serialVersionUID = 4806417891899193518L;
+
+ /**
+ * Constructor for MatrixNotSPDException
+ */
+ public MatrixNotSPDException() {
+ super();
+ }
+
+ /**
+ * Constructor for MatrixNotSPDException
+ *
+ * @param message
+ * Description of the exception
+ */
+ public MatrixNotSPDException(String message) {
+ super(message);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/MatrixSingularException.java b/src/main/java/no/uib/cipr/matrix/MatrixSingularException.java
new file mode 100644
index 0000000..d85d4b9
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/MatrixSingularException.java
@@ -0,0 +1,47 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Matrix is singular
+ */
+public class MatrixSingularException extends RuntimeException {
+
+ private static final long serialVersionUID = -8054618754675367225L;
+
+ /**
+ * Constructor for MatrixSingularException
+ */
+ public MatrixSingularException() {
+ super();
+ }
+
+ /**
+ * Constructor for MatrixSingularException
+ *
+ * @param message
+ * Description of the exception
+ */
+ public MatrixSingularException(String message) {
+ super(message);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/NotConvergedException.java b/src/main/java/no/uib/cipr/matrix/NotConvergedException.java
new file mode 100644
index 0000000..541210a
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/NotConvergedException.java
@@ -0,0 +1,85 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Signals lack of convergence of an iterative process
+ */
+public class NotConvergedException extends Exception {
+
+ private static final long serialVersionUID = -2305369220010776320L;
+
+ /**
+ * Possible reasons for lack of convergence
+ */
+ public enum Reason {
+
+ /**
+ * Did not converge after a maximum number of iterations
+ */
+ Iterations,
+
+ /**
+ * Divergence detected
+ */
+ Divergence,
+
+ /**
+ * The iterative process detected a breakdown
+ */
+ Breakdown
+ }
+
+ /**
+ * The reason for this exception
+ */
+ protected Reason reason;
+
+ /**
+ * Constructor for NotConvergedException
+ *
+ * @param reason
+ * The reason for the lack of convergence
+ * @param message
+ * A more descriptive explanation
+ */
+ public NotConvergedException(Reason reason, String message) {
+ super(message);
+ this.reason = reason;
+ }
+
+ /**
+ * Constructor for NotConvergedException. No message is provided
+ *
+ * @param reason
+ * The reason for the lack of convergence
+ */
+ public NotConvergedException(Reason reason) {
+ this.reason = reason;
+ }
+
+ /**
+ * Returns the reason for the exception
+ */
+ public Reason getReason() {
+ return reason;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/OrthogonalComputer.java b/src/main/java/no/uib/cipr/matrix/OrthogonalComputer.java
new file mode 100644
index 0000000..a0f26b6
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/OrthogonalComputer.java
@@ -0,0 +1,103 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Base class for the orthogonal matrix decompositions (QR, RQ, LQ, and QL)
+ */
+abstract class OrthogonalComputer {
+
+ /**
+ * The orthogonal matrix
+ */
+ final DenseMatrix Q;
+
+ /**
+ * Lower triangular factor. May not be present
+ */
+ final LowerTriangDenseMatrix L;
+
+ /**
+ * Upper triangular factor. May not be present
+ */
+ final UpperTriangDenseMatrix R;
+
+ /**
+ * Factorisation sizes
+ */
+ final int m, n, k;
+
+ /**
+ * Work arrays
+ */
+ double[] work, workGen;
+
+ /**
+ * Scales for the reflectors
+ */
+ final double[] tau;
+
+ /**
+ * Constructor for OrthogonalComputer
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns
+ * @param upper
+ * True for storing an upper triangular factor, false for a lower
+ * triangular factor
+ */
+ OrthogonalComputer(int m, int n, boolean upper) {
+ this.m = m;
+ this.n = n;
+ this.k = Math.min(m, n);
+
+ tau = new double[k];
+
+ Q = new DenseMatrix(m, n);
+ if (upper) {
+ R = new UpperTriangDenseMatrix(Math.min(m, n));
+ L = null;
+ } else {
+ L = new LowerTriangDenseMatrix(Math.min(m, n));
+ R = null;
+ }
+ }
+
+ /**
+ * Computes an orthogonal decomposition
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on exit. Pass a copy to avoid
+ * this
+ * @return The current decomposition
+ */
+ public abstract OrthogonalComputer factor(DenseMatrix A);
+
+ /**
+ * Returns the orthogonal part of the factorization
+ */
+ public DenseMatrix getQ() {
+ return Q;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/PackCholesky.java b/src/main/java/no/uib/cipr/matrix/PackCholesky.java
new file mode 100644
index 0000000..76942bc
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/PackCholesky.java
@@ -0,0 +1,233 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrix.Norm;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+/**
+ * Packed Cholesky decomposition
+ */
+public class PackCholesky {
+
+ /**
+ * Matrix dimension
+ */
+ private final int n;
+
+ /**
+ * Cholesky decomposition of a lower matrix
+ */
+ private LowerTriangPackMatrix Cl;
+
+ /**
+ * Cholesky decomposition of an upper matrix
+ */
+ private UpperTriangPackMatrix Cu;
+
+ /**
+ * If the matrix is SPD or not
+ */
+ private boolean notspd;
+
+ /**
+ * True for upper part, else false
+ */
+ private final boolean upper;
+
+ /**
+ * Constructor for DenseCholesky
+ *
+ * @param n
+ * Matrix size
+ * @param upper
+ * True for decomposing an upper symmetrical matrix, false for a
+ * lower symmetrical matrix
+ */
+ public PackCholesky(int n, boolean upper) {
+ this.n = n;
+ this.upper = upper;
+
+ if (upper)
+ Cu = new UpperTriangPackMatrix(n);
+ else
+ Cl = new LowerTriangPackMatrix(n);
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return The current decomposition
+ */
+ public static PackCholesky factorize(Matrix A) {
+ return new PackCholesky(A.numRows(), true)
+ .factor(new UpperSPDPackMatrix(A));
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public PackCholesky factor(LowerSPDPackMatrix A) {
+ if (upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for upper matrices");
+
+ return decompose(A);
+ }
+
+ /**
+ * Calculates a Cholesky decomposition
+ *
+ * @param A
+ * Matrix to decompose. Overwritten on return
+ * @return The current decomposition
+ */
+ public PackCholesky factor(UpperSPDPackMatrix A) {
+ if (!upper)
+ throw new IllegalArgumentException(
+ "Cholesky decomposition constructed for lower matrices");
+
+ return decompose(A);
+ }
+
+ private PackCholesky decompose(AbstractPackMatrix A) {
+ if (n != A.numRows())
+ throw new IllegalArgumentException("n != A.numRows()");
+
+ notspd = false;
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpptrf(UpLo.Upper.netlib(), A.numRows(),
+ A.getData(), info);
+ else
+ LAPACK.getInstance().dpptrf(UpLo.Lower.netlib(), A.numRows(),
+ A.getData(), info);
+
+ if (info.val > 0)
+ notspd = true;
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ if (upper)
+ Cu.set(A);
+ else
+ Cl.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns true if the matrix decomposed is symmetrical, positive definite
+ */
+ public boolean isSPD() {
+ return !notspd;
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a lower
+ * SPD matrix
+ */
+ public LowerTriangPackMatrix getL() {
+ if (!upper)
+ return Cl;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Returns the decomposition matrix. Only valid for decomposition of a upper
+ * SPD matrix
+ */
+ public UpperTriangPackMatrix getU() {
+ if (upper)
+ return Cu;
+ else
+ throw new UnsupportedOperationException();
+ }
+
+ /**
+ * Solves for <code>B</code>, overwriting it on return
+ */
+ public DenseMatrix solve(DenseMatrix B) throws MatrixNotSPDException {
+ if (notspd)
+ throw new MatrixNotSPDException();
+ if (B.numRows() != n)
+ throw new IllegalArgumentException("B.numRows() != n");
+
+ intW info = new intW(0);
+ if (upper)
+ LAPACK.getInstance().dpptrs(UpLo.Upper.netlib(), Cu.numRows(),
+ B.numColumns(), Cu.getData(), B.getData(), Matrices.ld(Cu.numRows()), info);
+ else
+ LAPACK.getInstance().dpptrs(UpLo.Lower.netlib(), Cl.numRows(),
+ B.numColumns(), Cl.getData(), B.getData(), Matrices.ld(Cl.numRows()), info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return B;
+ }
+
+ /**
+ * Computes the reciprocal condition number
+ *
+ * @param A
+ * The matrix this is a decomposition of
+ * @return The reciprocal condition number. Values close to unity indicate a
+ * well-conditioned system, while numbers close to zero do not.
+ */
+ public double rcond(Matrix A) {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+
+ double anorm = A.norm(Norm.One);
+
+ double[] work = new double[3 * n];
+ int[] iwork = new int[n];
+
+ intW info = new intW(0);
+ doubleW rcond = new doubleW(0);
+ if (upper)
+ LAPACK.getInstance().dppcon(UpLo.Upper.netlib(), n, Cu.getData(), anorm,
+ rcond, work, iwork, info);
+ else
+ LAPACK.getInstance().dppcon(UpLo.Lower.netlib(), n, Cl.getData(), anorm,
+ rcond, work, iwork, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return rcond.val;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/PermutationMatrix.java b/src/main/java/no/uib/cipr/matrix/PermutationMatrix.java
new file mode 100644
index 0000000..e6ebec7
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/PermutationMatrix.java
@@ -0,0 +1,111 @@
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+
+import java.util.BitSet;
+
+/**
+ * Matrix that represents a permutation of another matrix's
+ * rows / columns.
+ * <p>
+ * NOTE: the transpose of a permutation matrix is its inverse.
+ *
+ * @author Sam Halliday
+ */
+public class PermutationMatrix extends AbstractMatrix {
+
+ /**
+ * The sequential row permutations to perform, e.g. (2, 3, 3) means:
+ * permute row 1 with row 2, then permute row 2 with row 3,
+ * then permute row 3 with row 3 (i.e. do nothing).
+ * <p>
+ * Using this factory will ensure that LAPACK optimisations are
+ * available for multiplication operations.
+ *
+ * @param pivots using fortran (1-indexed) notation.
+ */
+ public static PermutationMatrix fromPartialPivots(int pivots[]) {
+ int[] permutations = new int[pivots.length];
+ for (int i = 0; i < pivots.length; i++) {
+ permutations[i] = i;
+ }
+
+ for (int i = 0; i < pivots.length; i++) {
+ int j = pivots[i] - 1;
+ if (j == i) continue;
+ int tmp = permutations[i];
+ permutations[i] = permutations[j];
+ permutations[j] = tmp;
+ }
+
+ return new PermutationMatrix(permutations, pivots);
+ }
+
+ private int[] permutations, pivots;
+
+ private boolean transposed;
+
+ // the instantaneous permutations to perform (zero-indexed)
+ // http://en.wikipedia.org/wiki/Permutation_matrix
+ public PermutationMatrix(int permutations[]) {
+ this(permutations, null);
+ }
+
+ // permutations - instantaneous (zero-indexed)
+ // pivots - sequential (fortran-indexed)
+ private PermutationMatrix(int permutations[], int pivots[]) {
+ super(permutations.length, permutations.length);
+ this.permutations = permutations;
+ BitSet bitset = new BitSet();
+ for (int i : permutations) {
+ if (bitset.get(i))
+ throw new IllegalArgumentException("non-unique permutations: " + i);
+ bitset.set(i);
+ }
+ this.pivots = pivots;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (!transposed && permutations[row] == column) return 1;
+ if (transposed && permutations[column] == row) return 1;
+ return 0;
+ }
+
+ @Override
+ public Matrix transpose() {
+ transposed = !transposed;
+ return this;
+ }
+
+ @Override
+ public Matrix mult(Matrix B, Matrix C) {
+ if (C instanceof DenseMatrix) return mult(B, (DenseMatrix) C);
+ return super.mult(B, C);
+ }
+
+ public Matrix mult(Matrix B, DenseMatrix C) {
+ if (pivots == null) return super.mult(B, C);
+ checkMultAdd(B, C);
+ C.set(B);
+
+ LAPACK.getInstance().dlaswp(C.numColumns(), C.getData(), Matrices.ld(C.numRows()), 1, pivots.length, pivots, transposed? -1 : 1);
+ return C;
+ }
+
+ @Override
+ public Matrix transAmult(Matrix B, Matrix C) {
+ if (C instanceof DenseMatrix) return transAmult(B, (DenseMatrix) C);
+ return super.transAmult(B, C);
+ }
+
+ public Matrix transAmult(Matrix B, DenseMatrix C) {
+ if (pivots == null) return super.transAmult(B, C);
+ checkTransAmultAdd(B, C);
+ C.set(B);
+
+ LAPACK.getInstance().dlaswp(C.numColumns(), C.getData(), Matrices.ld(C.numRows()), 1, pivots.length, pivots, transposed? 1 : -1);
+ return C;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/QL.java b/src/main/java/no/uib/cipr/matrix/QL.java
new file mode 100644
index 0000000..caa6cc9
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/QL.java
@@ -0,0 +1,139 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes QL decompositions
+ */
+public class QL extends OrthogonalComputer {
+
+ /**
+ * Constructs an empty QL decomposition
+ *
+ * @param m
+ * Number of rows. Must be larger than or equal the number of
+ * columns
+ * @param n
+ * Number of columns
+ */
+ public QL(int m, int n) {
+ super(m, n, false);
+
+ if (n > m)
+ throw new IllegalArgumentException("n > m");
+
+ int lwork;
+
+ // Query optimal workspace. First for computing the factorization
+ {
+ work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeqlf(m, n, new double[0], Matrices.ld(m), new double[0],
+ work, -1, info);
+
+ if (info.val != 0)
+ lwork = n;
+ else
+ lwork = (int) work[0];
+ lwork = Math.max(1, lwork);
+ work = new double[lwork];
+ }
+
+ // Workspace needed for generating an explicit orthogonal matrix
+ {
+ workGen = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dorgql(m, n, k, new double[0],
+ Matrices.ld(m), new double[0], workGen, -1, info);
+
+ if (info.val != 0)
+ lwork = n;
+ else
+ lwork = (int) workGen[0];
+ lwork = Math.max(1, lwork);
+ workGen = new double[lwork];
+ }
+
+ }
+
+ /**
+ * Convenience method to compute a QL decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return Newly allocated decomposition
+ */
+ public static QL factorize(Matrix A) {
+ return new QL(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ @Override
+ public QL factor(DenseMatrix A) {
+
+ if (Q.numRows() != A.numRows())
+ throw new IllegalArgumentException("Q.numRows() != A.numRows()");
+ else if (Q.numColumns() != A.numColumns())
+ throw new IllegalArgumentException(
+ "Q.numColumns() != A.numColumns()");
+ else if (L == null)
+ throw new IllegalArgumentException("L == null");
+
+ /*
+ * Calculate factorisation, and extract the triangular factor
+ */
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeqlf(m, n, A.getData(), Matrices.ld(m), tau, work,
+ work.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ L.zero();
+ for (MatrixEntry e : A)
+ if (e.row() >= (m - n) + e.column())
+ L.set(e.row() - (m - n), e.column(), e.get());
+
+ /*
+ * Generate the orthogonal matrix
+ */
+ info.val = 0;
+ LAPACK.getInstance().dorgql(m, n, k, A.getData(), Matrices.ld(m), tau, workGen,
+ workGen.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ Q.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the lower triangular factor
+ */
+ public LowerTriangDenseMatrix getL() {
+ return L;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/QR.java b/src/main/java/no/uib/cipr/matrix/QR.java
new file mode 100644
index 0000000..84d7a26
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/QR.java
@@ -0,0 +1,138 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computers QR decompositions
+ */
+public class QR extends OrthogonalComputer {
+
+ /**
+ * Constructs an empty QR decomposition
+ *
+ * @param m
+ * Number of rows. Must be larger than or equal the number of
+ * columns
+ * @param n
+ * Number of columns
+ */
+ public QR(int m, int n) {
+ super(m, n, true);
+
+ if (n > m)
+ throw new IllegalArgumentException("n > m");
+
+ int lwork;
+
+ // Query optimal workspace. First for computing the factorization
+ {
+ work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeqrf(m, n, new double[0], Matrices.ld(m), new double[0],
+ work, -1, info);
+
+ if (info.val != 0)
+ lwork = n;
+ else
+ lwork = (int) work[0];
+ lwork = Math.max(1, lwork);
+ work = new double[lwork];
+ }
+
+ // Workspace needed for generating an explicit orthogonal matrix
+ {
+ workGen = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dorgqr(m, n, k, new double[0],
+ Matrices.ld(m), new double[0], workGen, -1, info);
+
+ if (info.val != 0)
+ lwork = n;
+ else
+ lwork = (int) workGen[0];
+ lwork = Math.max(1, lwork);
+ workGen = new double[lwork];
+ }
+
+ }
+
+ /**
+ * Convenience method to compute a QR decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return Newly allocated decomposition
+ */
+ public static QR factorize(Matrix A) {
+ return new QR(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ @Override
+ public QR factor(DenseMatrix A) {
+
+ if (Q.numRows() != A.numRows())
+ throw new IllegalArgumentException("Q.numRows() != A.numRows()");
+ else if (Q.numColumns() != A.numColumns())
+ throw new IllegalArgumentException(
+ "Q.numColumns() != A.numColumns()");
+ else if (R == null)
+ throw new IllegalArgumentException("R == null");
+
+ /*
+ * Calculate factorisation, and extract the triangular factor
+ */
+ intW info = new intW(0);
+ LAPACK.getInstance().dgeqrf(m, n, A.getData(), Matrices.ld(m), tau, work,
+ work.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ R.zero();
+ for (MatrixEntry e : A)
+ if (e.row() <= e.column())
+ R.set(e.row(), e.column(), e.get());
+
+ /*
+ * Generate the orthogonal matrix
+ */
+ info.val = 0;
+ LAPACK.getInstance().dorgqr(m, n, k, A.getData(), Matrices.ld(m), tau, workGen,
+ workGen.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ Q.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the upper triangular factor
+ */
+ public UpperTriangDenseMatrix getR() {
+ return R;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/QRP.java b/src/main/java/no/uib/cipr/matrix/QRP.java
new file mode 100644
index 0000000..da83d50
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/QRP.java
@@ -0,0 +1,224 @@
+/*
+ * Copyright (C) 2006 Rafael de Pelegrini Soares
+ *
+ * MTJ additions.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes QR decompositions with column pivoting:
+ *
+ * {@code A*P = Q*R} where
+ *
+ * {@code A(m,n)}, {@code Q(m,m)}, and {@code R(m,n)}, more generally:
+ *
+ * {@code A*P = [Q1 Q2] * [R11, R12; 0 R22]} and {@code R22} elements are
+ * negligible.
+ *
+ */
+public class QRP {
+
+ /** Pivoting vector */
+ int jpvt[];
+ /**
+ * Scales for the reflectors
+ */
+ final double[] tau;
+ /**
+ * Factorisation sizes
+ */
+ final int m, n, k;
+ /** The factored matrix rank */
+ int rank;
+ /**
+ * Work array
+ */
+ double[] work;
+ /**
+ * The factored matrix
+ */
+ final DenseMatrix Afact;
+ /**
+ * The orthogonal matrix
+ */
+ final DenseMatrix Q;
+ /**
+ * The general upper triangular matrix.
+ */
+ final DenseMatrix R;
+
+ /**
+ * Constructs an empty QR decomposition
+ *
+ * @param m the number of rows.
+ * @param n the number of columns.
+ */
+ public QRP(int m, int n) {
+ this.m = m;
+ this.n = n;
+ this.k = Math.min(m, n);
+ this.rank = 0;
+ jpvt = new int[n];
+ tau = new double[k];
+
+ Q = new DenseMatrix(m, m);
+ R = new DenseMatrix(m, n);
+ Afact = new DenseMatrix(m, Math.max(m, n));
+
+ int lwork1, lwork2;
+ intW info = new intW(0);
+ double dummy[] = new double[1];
+ double ret[] = new double[1];
+
+ LAPACK lapack = LAPACK.getInstance();
+
+ // Query optimal workspace. First for computing the factorization
+ lapack.dgeqrf(m, n, dummy, Matrices.ld(m), dummy, ret, -1, info);
+ lwork1 = (info.val != 0) ? n : (int) ret[0];
+
+ // Workspace needed for generating an explicit orthogonal matrix
+ lapack.dorgqr(m, m, k, dummy, Matrices.ld(m), dummy, ret, -1, info);
+ lwork2 = (info.val != 0) ? n : (int) ret[0];
+
+ work = new double[Math.max(lwork1, lwork2)];
+ }
+
+ /**
+ * Convenience method to compute a QR decomposition
+ *
+ * @param A the matrix to decompose (not modified)
+ * @return Newly allocated decomposition
+ */
+ public static QRP factorize(Matrix A) {
+ return new QRP(A.numRows(), A.numColumns()).factor(A);
+ }
+
+ /**
+ * Executes a QR factorization for the given matrix.
+ *
+ * @param A the matrix to be factored (not modified)
+ * @return the factorization object
+ */
+ public QRP factor(Matrix A) {
+ if (Q.numRows() != A.numRows())
+ throw new IllegalArgumentException("Q.numRows() != A.numRows()");
+ else if (R.numColumns() != A.numColumns())
+ throw new IllegalArgumentException("R.numColumns() != A.numColumns()");
+
+ // copy A values in Afact
+ Afact.zero();
+ for (MatrixEntry e : A) {
+ Afact.set(e.row(), e.column(), e.get());
+ }
+
+ intW info = new intW(0);
+ LAPACK lapack = LAPACK.getInstance();
+
+ /*
+ * Calculate factorisation
+ */
+ lapack.dgeqp3(m, n, Afact.getData(), Matrices.ld(m), jpvt, tau, work,
+ work.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ /*
+ * Get R from Afact
+ */
+ R.zero();
+ for (MatrixEntry e : Afact) {
+ if (e.row() <= e.column() && e.column() < R.numColumns()) {
+ R.set(e.row(), e.column(), e.get());
+ }
+ }
+
+ /*
+ * Calculate the rank based on a precision EPS
+ */
+ final double EPS = 1e-12;
+ for (rank = 0; rank < k; rank++) {
+ if (Math.abs(R.get(rank, rank)) < EPS)
+ break;
+ }
+
+ /*
+ * Explicit the orthogonal matrix
+ */
+ lapack.dorgqr(m, m, k, Afact.getData(), Matrices.ld(m), tau, work,
+ work.length, info);
+ for (MatrixEntry e : Afact) {
+ if (e.column() < Q.numColumns())
+ Q.set(e.row(), e.column(), e.get());
+ }
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ // Adjust the permutation to zero offset
+ for (int i = 0; i < jpvt.length; i++) {
+ --jpvt[i];
+ }
+
+ return this;
+ }
+
+ /**
+ * Returns the upper triangular factor
+ */
+ public DenseMatrix getR() {
+ return R;
+ }
+
+ /**
+ * Returns the orthogonal matrix
+ */
+ public DenseMatrix getQ() {
+ return Q;
+ }
+
+ /**
+ * Returns the column pivoting vector.
+ * This function is cheaper than {@link #getP()}.
+ */
+ public int[] getPVector() {
+ return jpvt;
+ }
+
+ /**
+ * Returns the column pivoting matrix.
+ * This function allocates a new Matrix to be returned,
+ * a more cheap option is tu use {@link #getPVector()}.
+ */
+ public Matrix getP() {
+ Matrix P = new DenseMatrix(jpvt.length, jpvt.length);
+ for (int i = 0; i < jpvt.length; i++) {
+ P.set(jpvt[i], i, 1);
+ }
+ return P;
+ }
+
+ /**
+ * Returns the rank of the factored matrix
+ */
+ public int getRank() {
+ return rank;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/RQ.java b/src/main/java/no/uib/cipr/matrix/RQ.java
new file mode 100644
index 0000000..395f037
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/RQ.java
@@ -0,0 +1,138 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes RQ decompositions
+ */
+public class RQ extends OrthogonalComputer {
+
+ /**
+ * Constructs an empty RQ decomposition
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns. Must be larger than or equal the number of
+ * rows
+ */
+ public RQ(int m, int n) {
+ super(m, n, true);
+
+ if (n < m)
+ throw new IllegalArgumentException("n < m");
+
+ int lwork;
+
+ // Query optimal workspace. First for computing the factorization
+ {
+ work = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgerqf(m, n, new double[0], Matrices.ld(m), new double[0],
+ work, -1, info);
+
+ if (info.val != 0)
+ lwork = m;
+ else
+ lwork = (int) work[0];
+ lwork = Math.max(1, lwork);
+ work = new double[lwork];
+ }
+
+ // Workspace needed for generating an explicit orthogonal matrix
+ {
+ workGen = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dorgrq(m, n, m, new double[0],Matrices.ld(m),
+ new double[0], workGen, -1, info);
+
+ if (info.val != 0)
+ lwork = m;
+ else
+ lwork = (int) workGen[0];
+ lwork = Math.max(1, lwork);
+ workGen = new double[lwork];
+ }
+
+ }
+
+ /**
+ * Convenience method to compute an RQ decomposition
+ *
+ * @param A
+ * Matrix to decompose. Not modified
+ * @return Newly allocated decomposition
+ */
+ public static RQ factorize(Matrix A) {
+ return new RQ(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ @Override
+ public RQ factor(DenseMatrix A) {
+
+ if (Q.numRows() != A.numRows())
+ throw new IllegalArgumentException("Q.numRows() != A.numRows()");
+ else if (Q.numColumns() != A.numColumns())
+ throw new IllegalArgumentException(
+ "Q.numColumns() != A.numColumns()");
+ else if (R == null)
+ throw new IllegalArgumentException("R == null");
+
+ /*
+ * Calculate factorisation, and extract the triangular factor
+ */
+ intW info = new intW(0);
+ LAPACK.getInstance().dgerqf(m, n, A.getData(), Matrices.ld(m), tau, work,
+ work.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ R.zero();
+ for (MatrixEntry e : A)
+ if (e.column() >= (n - m) + e.row())
+ R.set(e.row(), e.column() - (n - m), e.get());
+
+ /*
+ * Generate the orthogonal matrix
+ */
+ info.val = 0;
+ LAPACK.getInstance().dorgrq(m, n, k, A.getData(), Matrices.ld(m), tau, workGen,
+ workGen.length, info);
+
+ if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ Q.set(A);
+
+ return this;
+ }
+
+ /**
+ * Returns the upper triangular factor
+ */
+ public UpperTriangDenseMatrix getR() {
+ return R;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SPDTridiagMatrix.java b/src/main/java/no/uib/cipr/matrix/SPDTridiagMatrix.java
new file mode 100644
index 0000000..f5558c7
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SPDTridiagMatrix.java
@@ -0,0 +1,97 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Symmetrical positive definite tridiagonal matrix. Same as
+ * {@link no.uib.cipr.matrix.SymmTridiagMatrix SymmTridiagMatrix}, and is used
+ * as a marker class to allow for more efficient solvers.
+ */
+public class SPDTridiagMatrix extends SymmTridiagMatrix {
+
+ /**
+ * Constructor for SPDTridiagMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public SPDTridiagMatrix(int n) {
+ super(n);
+ }
+
+ /**
+ * Constructor for SPDTridiagMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only main and the superdiagonal
+ * is copied over
+ */
+ public SPDTridiagMatrix(Matrix A) {
+ super(A);
+ }
+
+ /**
+ * Constructor for SPDTridiagMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only main and the superdiagonal
+ * is copied over
+ * @param deep
+ * True for a deep copy. For shallow copies <code>A</code> must
+ * be a <code>SymmTridiagMatrix</code>
+ */
+ public SPDTridiagMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ @Override
+ public SPDTridiagMatrix copy() {
+ return new SPDTridiagMatrix(this);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dptsv(numRows, X.numColumns(),
+ diag.clone(), offDiag.clone(), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixNotSPDException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SVD.java b/src/main/java/no/uib/cipr/matrix/SVD.java
new file mode 100644
index 0000000..d87fdd3
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SVD.java
@@ -0,0 +1,215 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes singular value decompositions
+ */
+public class SVD {
+
+ /**
+ * Work array
+ */
+ private final double[] work;
+
+ /**
+ * Work array
+ */
+ private final int[] iwork;
+
+ /**
+ * Matrix dimension
+ */
+ private final int m, n;
+
+ /**
+ * Compute the singular vectors fully?
+ */
+ private final boolean vectors;
+
+ /**
+ * Job to do
+ */
+ private final JobSVD job;
+
+ /**
+ * The singular values
+ */
+ private final double[] S;
+
+ /**
+ * Singular vectors
+ */
+ private final DenseMatrix U, Vt;
+
+ /**
+ * Creates an empty SVD which will compute all singular values and vectors
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns
+ */
+ public SVD(int m, int n) {
+ this(m, n, true);
+ }
+
+ /**
+ * Creates an empty SVD
+ *
+ * @param m
+ * Number of rows
+ * @param n
+ * Number of columns
+ * @param vectors
+ * True to compute the singular vectors, false for just the
+ * singular values
+ */
+ public SVD(int m, int n, boolean vectors) {
+ this.m = m;
+ this.n = n;
+ this.vectors = vectors;
+
+ // Allocate space for the decomposition
+ S = new double[Math.min(m, n)];
+ if (vectors) {
+ U = new DenseMatrix(m, m);
+ Vt = new DenseMatrix(n, n);
+ } else
+ U = Vt = null;
+
+ job = vectors ? JobSVD.All : JobSVD.None;
+
+ // Find workspace requirements
+ iwork = new int[8 * Math.min(m, n)];
+
+ // Query optimal workspace
+ double[] worksize = new double[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dgesdd(job.netlib(), m, n, new double[0],
+ Matrices.ld(m), new double[0], new double[0], Matrices.ld(m),
+ new double[0], Matrices.ld(n), worksize, -1,
+ iwork, info);
+
+ // Allocate workspace
+ int lwork = -1;
+ if (info.val != 0) {
+ if (vectors)
+ lwork = 3
+ * Math.min(m, n)
+ * Math.min(m, n)
+ + Math.max(Math.max(m, n), 4 * Math.min(m, n)
+ * Math.min(m, n) + 4 * Math.min(m, n));
+ else
+ lwork = 3
+ * Math.min(m, n)
+ * Math.min(m, n)
+ + Math.max(Math.max(m, n), 5 * Math.min(m, n)
+ * Math.min(m, n) + 4 * Math.min(m, n));
+ } else
+ lwork = (int) worksize[0];
+
+ lwork = Math.max(lwork, 1);
+ work = new double[lwork];
+ }
+
+ /**
+ * Convenience method for computing a full SVD
+ *
+ * @param A
+ * Matrix to decompose, not modified
+ * @return Newly allocated factorization
+ * @throws NotConvergedException
+ */
+ public static SVD factorize(Matrix A) throws NotConvergedException {
+ return new SVD(A.numRows(), A.numColumns()).factor(new DenseMatrix(A));
+ }
+
+ /**
+ * Computes an SVD
+ *
+ * @param A
+ * Matrix to decompose. Size must conform, and it will be
+ * overwritten on return. Pass a copy to avoid this
+ * @return The current decomposition
+ * @throws NotConvergedException
+ */
+ public SVD factor(DenseMatrix A) throws NotConvergedException {
+ if (A.numRows() != m)
+ throw new IllegalArgumentException("A.numRows() != m");
+ else if (A.numColumns() != n)
+ throw new IllegalArgumentException("A.numColumns() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgesdd(job.netlib(), m, n, A.getData(), Matrices.ld(m), S,
+ vectors ? U.getData() : new double[0], Matrices.ld(m),
+ vectors ? Vt.getData() : new double[0], Matrices.ld(n), work, work.length,
+ iwork, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+ /**
+ * True if singular vectors are stored
+ */
+ public boolean hasSingularVectors() {
+ return U != null;
+ }
+
+ /**
+ * Returns the left singular vectors, column-wise. Not available for partial
+ * decompositions
+ *
+ * @return Matrix of size m*m
+ */
+ public DenseMatrix getU() {
+ return U;
+ }
+
+ /**
+ * Returns the right singular vectors, row-wise. Not available for partial
+ * decompositions
+ *
+ * @return Matrix of size n*n
+ */
+ public DenseMatrix getVt() {
+ return Vt;
+ }
+
+ /**
+ * Returns the singular values (stored in descending order)
+ *
+ * @return Array of size min(m,n)
+ */
+ public double[] getS() {
+ return S;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Side.java b/src/main/java/no/uib/cipr/matrix/Side.java
new file mode 100644
index 0000000..98a2041
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Side.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/** Side enumeration */
+enum Side {
+ /** Apply operation from left */
+ Left,
+
+ /** Apply operation from right */
+ Right;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ if (this == Left)
+ return "L";
+ return "R";
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmBandEVD.java b/src/main/java/no/uib/cipr/matrix/SymmBandEVD.java
new file mode 100644
index 0000000..93299de
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmBandEVD.java
@@ -0,0 +1,177 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Computes eigenvalues of symmetrical, banded matrices
+ */
+public class SymmBandEVD extends SymmEVD {
+
+ /**
+ * Double work array
+ */
+ private final double[] work;
+
+ /**
+ * Integer work array
+ */
+ private final int[] iwork;
+
+ /**
+ * Upper or lower part stored
+ */
+ private final UpLo uplo;
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, banded matrices.
+ * Computes all eigenvalues and eigenvectors
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ */
+ public SymmBandEVD(int n, boolean upper) {
+ this(n, upper, true);
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, banded matrices
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ */
+ public SymmBandEVD(int n, boolean upper, boolean vectors) {
+ super(n, vectors);
+
+ uplo = upper ? UpLo.Upper : UpLo.Lower;
+
+ // Find the needed workspace
+ double[] worksize = new double[1];
+ int[] iworksize = new int[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dsbevd(job.netlib(), uplo.netlib(), n, 0, new double[0],
+ 1, new double[0], new double[0], Matrices.ld(n), worksize, -1, iworksize, -1, info);
+
+ // Allocate workspace
+ int lwork = 0, liwork = 0;
+ if (info.val != 0) {
+ if (job == JobEig.All) {
+ lwork = 1 + 6 * n + 2 * n * n;
+ liwork = 3 + 5 * n;
+ } else {
+ lwork = 2 * n;
+ liwork = 1;
+ }
+ } else {
+ lwork = (int) worksize[0];
+ liwork = iworksize[0];
+ }
+
+ lwork = Math.max(1, lwork);
+ liwork = Math.max(1, liwork);
+ work = new double[lwork];
+ iwork = new int[liwork];
+ }
+
+ /**
+ * Convenience method for computing the full eigenvalue decomposition of the
+ * given matrix
+ *
+ * @param A
+ * Matrix to factorize. The upper triangular part is extracted,
+ * and the matrix is not modified
+ * @param kd
+ * Number of diagonals to extract
+ * @return Newly allocated decomposition
+ * @throws NotConvergedException
+ */
+ public static SymmBandEVD factorize(Matrix A, int kd)
+ throws NotConvergedException {
+ return new SymmBandEVD(A.numRows(), true)
+ .factor(new UpperSymmBandMatrix(A, kd));
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmBandEVD factor(LowerSymmBandMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Lower)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for lower-symmetrical matrices");
+
+ return factor(A, A.getData(), A.numSubDiagonals());
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmBandEVD factor(UpperSymmBandMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Upper)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for upper-symmetrical matrices");
+
+ return factor(A, A.getData(), A.numSuperDiagonals());
+ }
+
+ private SymmBandEVD factor(Matrix A, double[] data, int kd)
+ throws NotConvergedException {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dsbevd(job.netlib(), uplo.netlib(), n, kd, data, Matrices.ld(kd + 1),
+ w, job == JobEig.All ? Z.getData() : new double[0], Matrices.ld(n), work,
+ work.length, iwork, iwork.length, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmDenseEVD.java b/src/main/java/no/uib/cipr/matrix/SymmDenseEVD.java
new file mode 100644
index 0000000..5cf7f03
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmDenseEVD.java
@@ -0,0 +1,225 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Computes eigenvalues of symmetrical, dense matrices
+ */
+public class SymmDenseEVD extends SymmEVD {
+
+ /**
+ * Double work array
+ */
+ private final double[] work;
+
+ /**
+ * Integer work array
+ */
+ private final int[] iwork;
+
+ /**
+ * Upper or lower part stored
+ */
+ private final UpLo uplo;
+
+ /**
+ * Range of eigenvalues to compute
+ */
+ private final JobEigRange range;
+
+ /**
+ * Eigenvector supports
+ */
+ private final int[] isuppz;
+
+ /**
+ * Tolerance criteria
+ */
+ private final double abstol;
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, dense matrices.
+ * Computes all eigenvalues and eigenvectors, and uses a low default
+ * tolerance criteria
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ */
+ public SymmDenseEVD(int n, boolean upper) {
+ this(n, upper, true, LAPACK.getInstance().dlamch("Safe minimum"));
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, dense matrices.
+ * Computes all eigenvalues and eigenvectors
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ * @param abstol
+ * Absolute tolerance criteria
+ */
+ public SymmDenseEVD(int n, boolean upper, double abstol) {
+ this(n, upper, true, abstol);
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, dense matrices. Uses
+ * a low default tolerance criteria
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ */
+ public SymmDenseEVD(int n, boolean upper, boolean vectors) {
+ this(n, upper, vectors, LAPACK.getInstance().dlamch("Safe minimum"));
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, dense matrices
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ * @param abstol
+ * Absolute tolerance criteria
+ */
+ public SymmDenseEVD(int n, boolean upper, boolean vectors, double abstol) {
+ super(n, vectors);
+ this.abstol = abstol;
+
+ uplo = upper ? UpLo.Upper : UpLo.Lower;
+ range = JobEigRange.All;
+ isuppz = new int[2 * Math.max(1, n)];
+
+ // Find the needed workspace
+ double[] worksize = new double[1];
+ int[] iworksize = new int[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dsyevr(job.netlib(), range.netlib(), uplo.netlib(), n,
+ new double[0], Matrices.ld(n), 0, 0, 0, 0, abstol, new intW(1), new double[0], new double[0],
+ Matrices.ld(n), isuppz, worksize, -1, iworksize, -1, info);
+
+ // Allocate workspace
+ int lwork = 0, liwork = 0;
+ if (info.val != 0) {
+ lwork = 26 * n;
+ liwork = 10 * n;
+ } else {
+ lwork = (int) worksize[0];
+ liwork = iworksize[0];
+ }
+
+ lwork = Math.max(1, lwork);
+ liwork = Math.max(1, liwork);
+ work = new double[lwork];
+ iwork = new int[liwork];
+ }
+
+ /**
+ * Convenience method for computing the full eigenvalue decomposition of the
+ * given matrix
+ *
+ * @param A
+ * Matrix to factorize. Upper part extracted, and the matrix is
+ * not modified
+ * @return Newly allocated decomposition
+ * @throws NotConvergedException
+ */
+ public static SymmDenseEVD factorize(Matrix A) throws NotConvergedException {
+ return new SymmDenseEVD(A.numRows(), true)
+ .factor(new UpperSymmDenseMatrix(A));
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmDenseEVD factor(LowerSymmDenseMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Lower)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for lower-symmetrical matrices");
+
+ return factor(A, A.getData());
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmDenseEVD factor(UpperSymmDenseMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Upper)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for upper-symmetrical matrices");
+
+ return factor(A, A.getData());
+ }
+
+ private SymmDenseEVD factor(Matrix A, double[] data)
+ throws NotConvergedException {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dsyevr(job.netlib(), range.netlib(), uplo.netlib(), n, data,
+ Matrices.ld(n), 0, 0, 0, 0, abstol, new intW(1), w,
+ job == JobEig.All ? Z.getData() : new double[0], Matrices.ld(n), isuppz, work,
+ work.length, iwork, iwork.length, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmEVD.java b/src/main/java/no/uib/cipr/matrix/SymmEVD.java
new file mode 100644
index 0000000..41a783d
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmEVD.java
@@ -0,0 +1,100 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Symmetric eigenvalue decomposition
+ */
+abstract class SymmEVD {
+
+ /**
+ * Size of the matrix
+ */
+ final int n;
+
+ /**
+ * The eigenvalues
+ */
+ final double[] w;
+
+ /**
+ * The eigenvectors stored columnwise
+ */
+ final DenseMatrix Z;
+
+ /**
+ * Job to do
+ */
+ final JobEig job;
+
+ /**
+ * Allocates storage for an eigenvalue computation
+ *
+ * @param n
+ * Size of the matrix
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ */
+ public SymmEVD(int n, boolean vectors) {
+ this.n = n;
+ w = new double[n];
+ job = vectors ? JobEig.All : JobEig.Eigenvalues;
+
+ if (vectors)
+ Z = new DenseMatrix(n, n);
+ else
+ Z = null;
+ }
+
+ /**
+ * Allocates storage for an eigenvalue computation. Includes eigenvectors
+ *
+ * @param n
+ * Size of the matrix
+ */
+ public SymmEVD(int n) {
+ this(n, true);
+ }
+
+ /**
+ * Gets the eigenvalues (stored in ascending order)
+ */
+ public double[] getEigenvalues() {
+ return w;
+ }
+
+ /**
+ * Gets the eigenvectors, if available
+ */
+ public DenseMatrix getEigenvectors() {
+ return Z;
+ }
+
+ /**
+ * True if the eigenvectors have been computed
+ */
+ public boolean hasEigenvectors() {
+ return Z != null;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmPackEVD.java b/src/main/java/no/uib/cipr/matrix/SymmPackEVD.java
new file mode 100644
index 0000000..74380b9
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmPackEVD.java
@@ -0,0 +1,174 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+
+/**
+ * Computes eigenvalues of symmetrical, packed matrices
+ */
+public class SymmPackEVD extends SymmEVD {
+
+ /**
+ * Double work array
+ */
+ private final double[] work;
+
+ /**
+ * Integer work array
+ */
+ private final int[] iwork;
+
+ /**
+ * Upper or lower part stored
+ */
+ private final UpLo uplo;
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, packed matrices.
+ * Computes all eigenvalues and eigenvectors
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ */
+ public SymmPackEVD(int n, boolean upper) {
+ this(n, upper, true);
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, packed matrices
+ *
+ * @param n
+ * Size of the matrix
+ * @param upper
+ * True if the upper part of the matrix is stored, and false if
+ * the lower part of the matrix is stored instead
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ */
+ public SymmPackEVD(int n, boolean upper, boolean vectors) {
+ super(n, vectors);
+
+ uplo = upper ? UpLo.Upper : UpLo.Lower;
+
+ // Find the needed workspace
+ double[] worksize = new double[1];
+ int[] iworksize = new int[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dspevd(job.netlib(), uplo.netlib(), n, new double[0],
+ new double[0], new double[0], Matrices.ld(n), worksize, -1, iworksize, -1, info);
+
+ // Allocate workspace
+ int lwork = 0, liwork = 0;
+ if (info.val != 0) {
+ if (job == JobEig.All) {
+ lwork = 1 + 6 * n + n * n;
+ liwork = 3 + 5 * n;
+ } else {
+ lwork = 2 * n;
+ liwork = 1;
+ }
+ } else {
+ lwork = (int) worksize[0];
+ liwork = iworksize[0];
+ }
+
+ lwork = Math.max(1, lwork);
+ liwork = Math.max(1, liwork);
+ work = new double[lwork];
+ iwork = new int[liwork];
+ }
+
+ /**
+ * Convenience method for computing the full eigenvalue decomposition of the
+ * given matrix
+ *
+ * @param A
+ * Matrix to factorize. Upper part extracted, and the matrix is
+ * not modified
+ * @return Newly allocated decomposition
+ * @throws NotConvergedException
+ */
+ public static SymmPackEVD factorize(Matrix A) throws NotConvergedException {
+ return new SymmPackEVD(A.numRows(), true)
+ .factor(new UpperSymmPackMatrix(A));
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmPackEVD factor(LowerSymmPackMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Lower)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for lower-symmetrical matrices");
+
+ return factor(A, A.getData());
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmPackEVD factor(UpperSymmPackMatrix A)
+ throws NotConvergedException {
+ if (uplo != UpLo.Upper)
+ throw new IllegalArgumentException(
+ "Eigenvalue computer configured for upper-symmetrical matrices");
+
+ return factor(A, A.getData());
+ }
+
+ private SymmPackEVD factor(Matrix A, double[] data)
+ throws NotConvergedException {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dspevd(job.netlib(), uplo.netlib(), n, data, w,
+ job == JobEig.All ? Z.getData() : new double[0], Matrices.ld(n), work,
+ work.length, iwork, iwork.length, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmTridiagEVD.java b/src/main/java/no/uib/cipr/matrix/SymmTridiagEVD.java
new file mode 100644
index 0000000..b32ec68
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmTridiagEVD.java
@@ -0,0 +1,180 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Computes eigenvalues of symmetrical, tridiagonal matrices
+ */
+public class SymmTridiagEVD extends SymmEVD {
+
+ /**
+ * Double work array
+ */
+ private final double[] work;
+
+ /**
+ * Integer work array
+ */
+ private final int[] iwork;
+
+ /**
+ * Range of eigenvalues to compute
+ */
+ private final JobEigRange range;
+
+ /**
+ * Eigenvector supports
+ */
+ private final int[] isuppz;
+
+ /**
+ * Tolerance criteria
+ */
+ private final double abstol;
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, tridiagonal
+ * matrices. Computes all eigenvalues and eigenvectors, and uses a low
+ * default tolerance criteria
+ *
+ * @param n
+ * Size of the matrix
+ */
+ public SymmTridiagEVD(int n) {
+ this(n, true);
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, tridiagonal
+ * matrices. Computes all eigenvalues and eigenvectors
+ *
+ * @param n
+ * Size of the matrix
+ * @param abstol
+ * Absolute tolerance criteria
+ */
+ public SymmTridiagEVD(int n, double abstol) {
+ this(n, true, abstol);
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, tridiagonal
+ * matrices. Uses a low default tolerance criteria
+ *
+ * @param n
+ * Size of the matrix
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ */
+ public SymmTridiagEVD(int n, boolean vectors) {
+ this(n, vectors, LAPACK.getInstance().dlamch("Safe minimum"));
+ }
+
+ /**
+ * Sets up an eigenvalue decomposition for symmetrical, tridiagonal matrices
+ *
+ * @param n
+ * Size of the matrix
+ * @param vectors
+ * True to compute the eigenvectors, false for just the
+ * eigenvalues
+ * @param abstol
+ * Absolute tolerance criteria
+ */
+ public SymmTridiagEVD(int n, boolean vectors, double abstol) {
+ super(n, vectors);
+ this.abstol = abstol;
+
+ range = JobEigRange.All;
+ isuppz = new int[2 * Math.max(1, n)];
+
+ // Find the needed workspace
+ double[] worksize = new double[1];
+ int[] iworksize = new int[1];
+ intW info = new intW(0);
+ LAPACK.getInstance().dstevr(job.netlib(), range.netlib(), n, new double[0],
+ new double[0], 0, 0, 0, 0, abstol, new intW(1), new double[0],
+ new double[0], Matrices.ld(n), isuppz, worksize, -1, iworksize, -1, info);
+
+ // Allocate workspace
+ int lwork = 0, liwork = 0;
+ if (info.val != 0) {
+ lwork = 20 * n;
+ liwork = 10 * n;
+ } else {
+ lwork = (int) worksize[0];
+ liwork = iworksize[0];
+ }
+
+ lwork = Math.max(1, lwork);
+ liwork = Math.max(1, liwork);
+ work = new double[lwork];
+ iwork = new int[liwork];
+ }
+
+ /**
+ * Convenience method for computing the full eigenvalue decomposition of the
+ * given matrix
+ *
+ * @param A
+ * Matrix to factorize. Main diagonal and superdiagonal is
+ * copied, and the matrix is not modified
+ * @return Newly allocated decomposition
+ * @throws NotConvergedException
+ */
+ public static SymmTridiagEVD factorize(Matrix A)
+ throws NotConvergedException {
+ return new SymmTridiagEVD(A.numRows()).factor(new SymmTridiagMatrix(A));
+ }
+
+ /**
+ * Computes the eigenvalue decomposition of the given matrix
+ *
+ * @param A
+ * Matrix to factorize. Overwritten on return
+ * @return The current eigenvalue decomposition
+ * @throws NotConvergedException
+ */
+ public SymmTridiagEVD factor(SymmTridiagMatrix A)
+ throws NotConvergedException {
+ if (A.numRows() != n)
+ throw new IllegalArgumentException("A.numRows() != n");
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dstevr(job.netlib(), range.netlib(), n, A.getDiagonal(),
+ A.getOffDiagonal(), 0, 0, 0, 0, abstol, new intW(1), w,
+ job == JobEig.All ? Z.getData() : new double[0], Matrices.ld(n), isuppz, work,
+ work.length, iwork, iwork.length, info);
+
+ if (info.val > 0)
+ throw new NotConvergedException(
+ NotConvergedException.Reason.Iterations);
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return this;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/SymmTridiagMatrix.java b/src/main/java/no/uib/cipr/matrix/SymmTridiagMatrix.java
new file mode 100644
index 0000000..b26770b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/SymmTridiagMatrix.java
@@ -0,0 +1,324 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Symmetrical tridiagonal matrix. Storage as for
+ * {@link no.uib.cipr.matrix.TridiagMatrix TridiagMatrix}, but only one
+ * off-diagonal array is stored.
+ */
+public class SymmTridiagMatrix extends AbstractMatrix {
+
+ /**
+ * Diagonal and off-diagonal
+ */
+ double[] diag, offDiag;
+
+ /**
+ * Size of the matrix
+ */
+ int n;
+
+ /**
+ * Constructor for SymmTridiagMatrix
+ *
+ * @param diag
+ * Main diagonal
+ * @param offDiag
+ * Offdiagonals, both upper and lower
+ * @param n
+ * Size of the matrix. The main diagonal must be at least as long
+ * as n, and the off diagonal part must be at least as long as
+ * n-1
+ */
+ public SymmTridiagMatrix(double[] diag, double[] offDiag, int n) {
+ super(n, n);
+
+ this.n = n;
+ if (n < 1)
+ throw new IllegalArgumentException("n must be >= 1");
+
+ if (diag.length < n)
+ throw new IllegalArgumentException("diag.length < n");
+ if (offDiag.length < n - 1)
+ throw new IllegalArgumentException("offDiag.length < n - 1");
+
+ this.diag = diag;
+ this.offDiag = offDiag;
+ }
+
+ /**
+ * Constructor for SymmTridiagMatrix
+ *
+ * @param diag
+ * Main diagonal
+ * @param offDiag
+ * Offdiagonals. Must be one shorter than diag
+ */
+ public SymmTridiagMatrix(double[] diag, double[] offDiag) {
+ this(diag, offDiag, diag.length);
+ }
+
+ /**
+ * Constructor for SymmTridiagMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns. <code>n</code>
+ * cannot be zero
+ */
+ public SymmTridiagMatrix(int n) {
+ super(n, n);
+
+ if (n < 1)
+ throw new IllegalArgumentException("n must be >= 1");
+
+ this.n = numRows;
+ diag = new double[n];
+ offDiag = new double[n - 1];
+ }
+
+ /**
+ * Constructor for SymmTridiagMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only main and the superdiagonal
+ * is copied over
+ */
+ public SymmTridiagMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for SymmTridiagMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only main and the superdiagonal
+ * is copied over. It must be square and cannot have any zero
+ * dimension lengths
+ * @param deep
+ * True for a deep copy. For shallow copies <code>A</code> must
+ * be a <code>SymmTridiagMatrix</code>
+ */
+ public SymmTridiagMatrix(Matrix A, boolean deep) {
+ super(A);
+
+ if (!isSquare())
+ throw new IllegalArgumentException(
+ "Symmetric matrix must be square");
+ if (A.numRows() < 1)
+ throw new IllegalArgumentException("numRows must be >= 1");
+ n = numRows;
+
+ if (deep) {
+ diag = new double[n];
+ offDiag = new double[Math.max(n - 1, 0)];
+ for (MatrixEntry e : A)
+ if (e.row() == e.column() || e.row() == e.column() + 1)
+ set(e.row(), e.column(), e.get());
+ } else {
+ SymmTridiagMatrix B = (SymmTridiagMatrix) A;
+ this.diag = B.getDiagonal();
+ this.offDiag = B.getOffDiagonal();
+ }
+ }
+
+ /**
+ * Returns the diagonal entries. Length equal <code>n</code>
+ */
+ public double[] getDiagonal() {
+ return diag;
+ }
+
+ /**
+ * Returns the off diagonal entries. Length equal <code>n-1</code>
+ */
+ public double[] getOffDiagonal() {
+ return offDiag;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ check(row, column);
+ if (row == column)
+ diag[row] += value;
+ else if (row == column + 1)
+ offDiag[column] += value;
+ else if (row != column - 1)
+ throw new IndexOutOfBoundsException(
+ "Insertion index outside of band");
+ }
+
+ @Override
+ public double get(int row, int column) {
+ check(row, column);
+ if (row == column)
+ return diag[row];
+ else if (row == column + 1)
+ return offDiag[column];
+ else if (row == column - 1)
+ return offDiag[row];
+ else
+ return 0;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+ if (row == column)
+ diag[row] = value;
+ else if (row == column + 1)
+ offDiag[column] = value;
+ else if (row != column - 1)
+ throw new IndexOutOfBoundsException(
+ "Insertion index outside of band");
+ }
+
+ @Override
+ public SymmTridiagMatrix copy() {
+ return new SymmTridiagMatrix(this);
+ }
+
+ @Override
+ public SymmTridiagMatrix zero() {
+ Arrays.fill(diag, 0);
+ Arrays.fill(offDiag, 0);
+ return this;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+ intW info = new intW(0);
+ LAPACK.getInstance().dgtsv(numRows, X.numColumns(),
+ offDiag.clone(), diag.clone(), offDiag.clone(), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transSolve(Matrix B, Matrix X) {
+ return solve(B, X);
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ return solve(b, x);
+ }
+
+ @Override
+ public Matrix transpose() {
+ return this;
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new SymmTridiagMatrixIterator();
+ }
+
+ /**
+ * Iterator over a symmetrical, tridiagonal matrix
+ */
+ private class SymmTridiagMatrixIterator extends RefMatrixIterator {
+
+ /**
+ * Current band, starting with the main diagonal
+ */
+ private double[] band = diag;
+
+ /**
+ * Band index
+ */
+ private int bandIndex;
+
+ /**
+ * Which band in use (0 for main, 1 for off)
+ */
+ private int whichBand;
+
+ @Override
+ public boolean hasNext() {
+ return whichBand < 3;
+ }
+
+ @Override
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ // Move in the band
+ if (bandIndex < band.length - 1)
+ bandIndex++;
+ else {
+ // Move to the off-diagonal (twice)
+ bandIndex = 0;
+ whichBand++;
+
+ band = offDiag;
+
+ // If the off-diagonals are zero-sized, we are done
+ // This happens if the matrix is 1*1
+ if (band.length == 0)
+ whichBand = 3;
+ }
+
+ // Set row index
+ if (whichBand == 1)
+ row = bandIndex + 1;
+ else
+ row = bandIndex;
+
+ // Set column index
+ if (whichBand == 2)
+ column = bandIndex + 1;
+ else
+ column = bandIndex;
+
+ return entry;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Transpose.java b/src/main/java/no/uib/cipr/matrix/Transpose.java
new file mode 100644
index 0000000..af3e7e0
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Transpose.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/** Transpose enumeration */
+enum Transpose {
+ /** Do not transpose */
+ NoTranspose,
+
+ /** Transpose */
+ Transpose;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ if (this == NoTranspose)
+ return "N";
+ return "T";
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/TridiagMatrix.java b/src/main/java/no/uib/cipr/matrix/TridiagMatrix.java
new file mode 100644
index 0000000..1d2799e
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/TridiagMatrix.java
@@ -0,0 +1,293 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+import com.github.fommil.netlib.LAPACK;
+import org.netlib.util.intW;
+
+/**
+ * Tridiagonal matrix. Stored in three arrays, one of length <code>n</code>
+ * for the diagonal, two of length <code>n-1</code> for the superdiagonal and
+ * subdiagonal entries.
+ */
+public class TridiagMatrix extends AbstractMatrix {
+
+ /**
+ * Diagonal, super-diagonal and sub-diagonal
+ */
+ double[] diag, superDiag, subDiag;
+
+ /**
+ * Size of the matrix
+ */
+ private int n;
+
+ /**
+ * Constructor for TridiagMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public TridiagMatrix(int n) {
+ super(n, n);
+
+ if (n < 1)
+ throw new IllegalArgumentException("n must be >= 1");
+
+ this.n = n;
+ diag = new double[n];
+ superDiag = new double[n - 1];
+ subDiag = new double[n - 1];
+ }
+
+ /**
+ * Constructor for TridiagMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the tridiagonal part is copied
+ */
+ public TridiagMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for TridiagMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the tridiagonal part is copied
+ * @param deep
+ * True for a deep copy, else it's shallow. For shallow copies,
+ * <code>A</code> must be a <code>TridiagMatrix</code>
+ */
+ public TridiagMatrix(Matrix A, boolean deep) {
+ super(A);
+
+ if (!isSquare())
+ throw new IllegalArgumentException(
+ "Tridiagonal matrix must be square");
+ if (A.numRows() < 1)
+ throw new IllegalArgumentException("numRows must be >= 1");
+ n = numRows;
+
+ if (deep) {
+ diag = new double[n];
+ superDiag = new double[n - 1];
+ subDiag = new double[n - 1];
+ for (MatrixEntry e : A)
+ if (e.row() == e.column() || e.row() == e.column() - 1
+ || e.row() == e.column() + 1)
+ set(e.row(), e.column(), e.get());
+ } else {
+ TridiagMatrix B = (TridiagMatrix) A;
+ this.diag = B.getDiagonal();
+ this.subDiag = B.getSubDiagonal();
+ this.superDiag = B.getSuperDiagonal();
+ }
+ }
+
+ /**
+ * Returns the diagonal entries. Length equal <code>n</code>
+ */
+ public double[] getDiagonal() {
+ return diag;
+ }
+
+ /**
+ * Returns the sub diagonal entries. Length equal <code>n-1</code>
+ */
+ public double[] getSubDiagonal() {
+ return subDiag;
+ }
+
+ /**
+ * Returns the super diagonal entries. Length equal <code>n-1</code>
+ */
+ public double[] getSuperDiagonal() {
+ return superDiag;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ check(row, column);
+ if (row == column)
+ diag[row] += value;
+ else if (row == column + 1)
+ subDiag[column] += value;
+ else if (row == column - 1)
+ superDiag[row] += value;
+ else
+ throw new IndexOutOfBoundsException(
+ "Insertion index outside of band");
+ }
+
+ @Override
+ public double get(int row, int column) {
+ check(row, column);
+ if (row == column)
+ return diag[row];
+ else if (row == column + 1)
+ return subDiag[column];
+ else if (row == column - 1)
+ return superDiag[row];
+ else
+ return 0;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+ if (row == column)
+ diag[row] = value;
+ else if (row == column + 1)
+ subDiag[column] = value;
+ else if (row == column - 1)
+ superDiag[row] = value;
+ else
+ throw new IndexOutOfBoundsException(
+ "Insertion index outside of band");
+ }
+
+ @Override
+ public TridiagMatrix copy() {
+ return new TridiagMatrix(this);
+ }
+
+ @Override
+ public TridiagMatrix zero() {
+ Arrays.fill(diag, 0);
+ Arrays.fill(subDiag, 0);
+ Arrays.fill(superDiag, 0);
+ return this;
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ if (!(X instanceof DenseMatrix))
+ throw new UnsupportedOperationException("X must be a DenseMatrix");
+
+ checkSolve(B, X);
+
+ double[] Xd = ((DenseMatrix) X).getData();
+
+ X.set(B);
+
+ intW info = new intW(0);
+ LAPACK.getInstance().dgtsv(numRows, X.numColumns(),
+ subDiag.clone(), diag.clone(), superDiag.clone(), Xd, Matrices.ld(numRows), info);
+
+ if (info.val > 0)
+ throw new MatrixSingularException();
+ else if (info.val < 0)
+ throw new IllegalArgumentException();
+
+ return X;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ DenseMatrix B = new DenseMatrix(b, false), X = new DenseMatrix(x, false);
+ solve(B, X);
+ return x;
+ }
+
+ @Override
+ public Matrix transpose() {
+ double[] otherDiag = subDiag;
+ subDiag = superDiag;
+ superDiag = otherDiag;
+ return this;
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new TridiagMatrixIterator();
+ }
+
+ /**
+ * Iterator over a tridiagonal matrix
+ */
+ private class TridiagMatrixIterator extends RefMatrixIterator {
+
+ /**
+ * Current band, starting with the main diagonal
+ */
+ private double[] band = diag;
+
+ /**
+ * Band index
+ */
+ private int bandIndex;
+
+ /**
+ * Which band in use (0 for main, 1 for sub, 2 for super)
+ */
+ private int whichBand;
+
+ @Override
+ public boolean hasNext() {
+ return whichBand < 3;
+ }
+
+ @Override
+ public MatrixEntry next() {
+ entry.update(row, column);
+
+ // Move in the band
+ if (bandIndex < band.length - 1)
+ bandIndex++;
+ else {
+ // Move to the next band
+ bandIndex = 0;
+ whichBand++;
+
+ if (whichBand == 1)
+ band = subDiag;
+ else if (whichBand == 2)
+ band = superDiag;
+
+ // If the off-diagonals are zero-sized, we are done
+ // This happens if the matrix is 1*1
+ if (band.length == 0)
+ whichBand = 3;
+ }
+
+ // Set row index
+ if (whichBand == 1)
+ row = bandIndex + 1;
+ else
+ row = bandIndex;
+
+ // Set column index
+ if (whichBand == 2)
+ column = bandIndex + 1;
+ else
+ column = bandIndex;
+
+ return entry;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrix.java
new file mode 100644
index 0000000..8baacb6
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrix.java
@@ -0,0 +1,113 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit lower triangular banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.LowerTriangBandMatrix LowerTriangBandMatrix}, but
+ * the main diagonal is assumed to be all ones. It is still allocated, however.
+ */
+public class UnitLowerTriangBandMatrix extends LowerTriangBandMatrix {
+
+ /**
+ * Constructor for UnitLowerTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public UnitLowerTriangBandMatrix(int n, int kd) {
+ super(n, kd, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored (including main diagonal entries)
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ */
+ public UnitLowerTriangBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored (including main diagonal entries)
+ * @param kd
+ * Number of bands below the main diagonal (subdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public UnitLowerTriangBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row == column)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.set(row, column, value);
+ }
+
+ @Override
+ public UnitLowerTriangBandMatrix copy() {
+ return new UnitLowerTriangBandMatrix(this, kl);
+ }
+
+ @Override
+ public Matrix zero() {
+ throw new UnsupportedOperationException();
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (inBand(e.row(), e.column()) && e.row() != e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrix.java
new file mode 100644
index 0000000..2544568
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrix.java
@@ -0,0 +1,107 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit lower triangular dense matrix. Almost the same as the
+ * {@link no.uib.cipr.matrix.LowerTriangDenseMatrix LowerTriangDenseMatrix},
+ * but additionally assumes the main diagonal to be all ones. However it does
+ * not access it, so it may be actually be different.
+ */
+public class UnitLowerTriangDenseMatrix extends LowerTriangDenseMatrix {
+
+ /**
+ * Constructor for UnitLowerTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UnitLowerTriangDenseMatrix(int n) {
+ super(n, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the strictly lower triangular part
+ * is copied
+ */
+ public UnitLowerTriangDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the strictly lower triangular part
+ * is copied
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ public UnitLowerTriangDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column == row)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.set(row, column, value);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() > e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UnitLowerTriangDenseMatrix copy() {
+ return new UnitLowerTriangDenseMatrix(this);
+ }
+
+ @Override
+ public Matrix zero() {
+ throw new UnsupportedOperationException();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrix.java
new file mode 100644
index 0000000..1f36939
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrix.java
@@ -0,0 +1,100 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit lower triangular packed matrix. Same storage as
+ * {@link no.uib.cipr.matrix.LowerTriangPackMatrix LowerTriangPackMatrix}, but
+ * the main diagonal is assumed to be all ones.
+ */
+public class UnitLowerTriangPackMatrix extends LowerTriangPackMatrix {
+
+ /**
+ * Constructor for UnitLowerTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UnitLowerTriangPackMatrix(int n) {
+ super(n, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public UnitLowerTriangPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UnitLowerTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public UnitLowerTriangPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (column == row)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.set(row, column, value);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() > e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UnitLowerTriangPackMatrix copy() {
+ return new UnitLowerTriangPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrix.java
new file mode 100644
index 0000000..9fc605c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrix.java
@@ -0,0 +1,113 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit upper triangular banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.LowerTriangBandMatrix LowerTriangBandMatrix}, but
+ * the main diagonal is assumed to be all ones. It is still allocated, however.
+ */
+public class UnitUpperTriangBandMatrix extends UpperTriangBandMatrix {
+
+ /**
+ * Constructor for UnitUpperTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ */
+ public UnitUpperTriangBandMatrix(int n, int kd) {
+ super(n, kd, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored (including main diagonal entries)
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ */
+ public UnitUpperTriangBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored (including main diagonal entries)
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public UnitUpperTriangBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row == column)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.set(row, column, value);
+ }
+
+ @Override
+ public UnitUpperTriangBandMatrix copy() {
+ return new UnitUpperTriangBandMatrix(this, ku);
+ }
+
+ @Override
+ public Matrix zero() {
+ throw new UnsupportedOperationException();
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (inBand(e.row(), e.column()) && e.row() != e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrix.java
new file mode 100644
index 0000000..9a20dae
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrix.java
@@ -0,0 +1,109 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit upper triangular dense matrix. Almost the same as the
+ * {@link no.uib.cipr.matrix.UpperTriangDenseMatrix UpperTriangDenseMatrix},
+ * but additionally assumes the main diagonal to be all ones. However it does
+ * not access it, so it may be actually be different.
+ */
+public class UnitUpperTriangDenseMatrix extends UpperTriangDenseMatrix {
+
+ /**
+ * Constructor for UnitUpperTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UnitUpperTriangDenseMatrix(int n) {
+ super(n, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangDenseMatrix. Matrix is copied from the
+ * supplied matrix
+ *
+ * @param A
+ * Matrix to copy from. Only the strictly upper triangular part
+ * is copied
+ */
+ public UnitUpperTriangDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangDenseMatrix. Matrix is copied from the
+ * supplied matrix
+ *
+ * @param A
+ * Matrix to copy from. Only the strictly upper triangular part
+ * is copied
+ * @param deep
+ * If true, <code>A</code> is copied, else a shallow copy is
+ * made and the matrices share underlying storage. For this,
+ * <code>A</code> must be a dense matrix
+ */
+ public UnitUpperTriangDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row == column)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (column == row)
+ throw new IllegalArgumentException("column == row");
+ super.set(row, column, value);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() < e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UnitUpperTriangDenseMatrix copy() {
+ return new UnitUpperTriangDenseMatrix(this);
+ }
+
+ @Override
+ public Matrix zero() {
+ throw new UnsupportedOperationException();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrix.java b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrix.java
new file mode 100644
index 0000000..015a29f
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrix.java
@@ -0,0 +1,100 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Unit upper triangular packed matrix. Same storage as
+ * {@link no.uib.cipr.matrix.UpperTriangPackMatrix UpperTriangPackMatrix}, but
+ * the main diagonal is assumed to be all ones.
+ */
+public class UnitUpperTriangPackMatrix extends UpperTriangPackMatrix {
+
+ /**
+ * Constructor for UnitUpperTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UnitUpperTriangPackMatrix(int n) {
+ super(n, Diag.Unit);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public UnitUpperTriangPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UnitUpperTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public UnitUpperTriangPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, Diag.Unit);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row == column)
+ return 1;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row == column)
+ throw new IllegalArgumentException("row == column");
+ super.set(row, column, value);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() < e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UnitUpperTriangPackMatrix copy() {
+ return new UnitUpperTriangPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpLo.java b/src/main/java/no/uib/cipr/matrix/UpLo.java
new file mode 100644
index 0000000..5f2754e
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpLo.java
@@ -0,0 +1,37 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+/**
+ * Designates the Upper or lower part of a Matrix.
+ */
+enum UpLo {
+ Upper, Lower;
+
+ /**
+ * @return the netlib character version of this designation, for use with F2J.
+ */
+ public String netlib() {
+ if (this == Upper)
+ return "U";
+ return "L";
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSPDBandMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSPDBandMatrix.java
new file mode 100644
index 0000000..ac0b604
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSPDBandMatrix.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Upper symmetrical positive definite banded matrix. It does not enforce this
+ * property (except for symmetry), and has the same storage layout as
+ * {@link no.uib.cipr.matrix.UpperSymmBandMatrix UpperSymmBandMatrix}.
+ */
+public class UpperSPDBandMatrix extends UpperSymmBandMatrix {
+
+ /**
+ * Constructor for UpperSPDBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public UpperSPDBandMatrix(int n, int kd) {
+ super(n, kd);
+ }
+
+ /**
+ * Constructor for UpperSPDBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public UpperSPDBandMatrix(Matrix A, int kd) {
+ super(A, kd);
+ }
+
+ /**
+ * Constructor for UpperSPDBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public UpperSPDBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, kd, deep);
+ }
+
+ @Override
+ public UpperSPDBandMatrix copy() {
+ return new UpperSPDBandMatrix(this, kd);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSPDDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSPDDenseMatrix.java
new file mode 100644
index 0000000..1d7b0dc
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSPDDenseMatrix.java
@@ -0,0 +1,77 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Upper symmetrical positive definite dense matrix. Same layout as
+ * {@link no.uib.cipr.matrix.UpperSymmDenseMatrix UpperSymmDenseMatrix}. This
+ * class does not enforce the SPD property, but serves as a tag so that more
+ * efficient algorithms can be used in the solvers.
+ */
+public class UpperSPDDenseMatrix extends UpperSymmDenseMatrix {
+
+ /**
+ * Constructor for UpperSPDDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperSPDDenseMatrix(int n) {
+ super(n);
+ }
+
+ /**
+ * Constructor for UpperSPDDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the upper
+ * triangular part is copied
+ */
+ public UpperSPDDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UpperSPDDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the upper
+ * triangular part is copied
+ * @param deep
+ * True for a deep copy, else shallow. In that case,
+ * <code>A</code> must be a dense matrix
+ */
+ public UpperSPDDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ @Override
+ public UpperSPDDenseMatrix copy() {
+ return new UpperSPDDenseMatrix(this);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSPDPackMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSPDPackMatrix.java
new file mode 100644
index 0000000..62bf871
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSPDPackMatrix.java
@@ -0,0 +1,77 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Upper symmetrical positive definite packed matrix. Same layout as
+ * {@link no.uib.cipr.matrix.UpperSymmPackMatrix UpperSymmPackMatrix}. This
+ * class does not enforce the SPD property, but serves as a tag so that more
+ * efficient algorithms can be used in the solvers.
+ */
+public class UpperSPDPackMatrix extends UpperSymmPackMatrix {
+
+ /**
+ * Constructor for UpperSPDPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperSPDPackMatrix(int n) {
+ super(n);
+ }
+
+ /**
+ * Constructor for UpperSPDPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public UpperSPDPackMatrix(Matrix A) {
+ super(A);
+ }
+
+ /**
+ * Constructor for UpperSPDPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public UpperSPDPackMatrix(Matrix A, boolean deep) {
+ super(A, deep);
+ }
+
+ @Override
+ public UpperSPDPackMatrix copy() {
+ return new UpperSPDPackMatrix(this);
+ }
+
+ @Override
+ public Matrix solve(Matrix B, Matrix X) {
+ return SPDsolve(B, X);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSymmBandMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSymmBandMatrix.java
new file mode 100644
index 0000000..f98c1c8
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSymmBandMatrix.java
@@ -0,0 +1,99 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper symmetrical banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.BandMatrix BandMatrix}, but without subdiagonals.
+ * Lower part of the matrix is implictly known by symmetry
+ */
+public class UpperSymmBandMatrix extends AbstractSymmBandMatrix {
+
+ /**
+ * Constructor for UpperSymmBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public UpperSymmBandMatrix(int n, int kd) {
+ super(n, 0, kd, UpLo.Upper);
+ }
+
+ /**
+ * Constructor for UpperSymmBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ */
+ public UpperSymmBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for UpperSymmBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands off the main diagonal (off diagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public UpperSymmBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, 0, kd, deep, UpLo.Upper);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row <= column)
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row > column)
+ return super.get(column, row);
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row <= column)
+ super.set(row, column, value);
+ }
+
+ @Override
+ public UpperSymmBandMatrix copy() {
+ return new UpperSymmBandMatrix(this, kd);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSymmDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSymmDenseMatrix.java
new file mode 100644
index 0000000..a3ed69f
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSymmDenseMatrix.java
@@ -0,0 +1,99 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper symmetrix dense matrix. It has the same storage layout as the
+ * {@link no.uib.cipr.matrix.DenseMatrix DenseMatrix}, but only refers to
+ * elements above or on the main diagonal. The remaining elements are never
+ * accessed nor changed, and is known only by symmetry.
+ */
+public class UpperSymmDenseMatrix extends AbstractSymmDenseMatrix {
+
+ /**
+ * Constructor for UpperSymmDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperSymmDenseMatrix(int n) {
+ super(n, UpLo.Upper);
+ }
+
+ /**
+ * Constructor for UpperSymmDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the upper
+ * triangular part is copied
+ */
+ public UpperSymmDenseMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UpperSymmDenseMatrix
+ *
+ * @param A
+ * Matrix to copy. It must be a square matrix, and only the upper
+ * triangular part is copied
+ * @param deep
+ * If false, a shallow copy is made. In that case, <code>A</code>
+ * must be a dense matrix
+ */
+ public UpperSymmDenseMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Upper);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row <= column)
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row > column)
+ return super.get(column, row);
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row <= column)
+ super.set(row, column, value);
+ }
+
+ @Override
+ public UpperSymmDenseMatrix copy() {
+ return new UpperSymmDenseMatrix(this);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() <= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperSymmPackMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperSymmPackMatrix.java
new file mode 100644
index 0000000..b941c36
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperSymmPackMatrix.java
@@ -0,0 +1,106 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper symmetric packed matrix. Same storage as
+ * {@link no.uib.cipr.matrix.UpperTriangPackMatrix UpperTriangPackMatrix}, but
+ * the lower triangular part is known by symmetry.
+ */
+public class UpperSymmPackMatrix extends AbstractSymmPackMatrix {
+
+ /**
+ * Constructor for UpperSymmPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperSymmPackMatrix(int n) {
+ super(n, UpLo.Upper);
+ }
+
+ /**
+ * Constructor for UpperSymmPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public UpperSymmPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UpperSymmPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public UpperSymmPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Upper);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row <= column)
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row <= column)
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row <= column)
+ return data[getIndex(row, column)];
+ return data[getIndex(column, row)];
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + (column + 1) * column / 2;
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() <= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UpperSymmPackMatrix copy() {
+ return new UpperSymmPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperTriangBandMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperTriangBandMatrix.java
new file mode 100644
index 0000000..0404752
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperTriangBandMatrix.java
@@ -0,0 +1,109 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper triangular banded matrix. The same storage as
+ * {@link no.uib.cipr.matrix.BandMatrix BandMatrix}, but without subdiagonals.
+ */
+public class UpperTriangBandMatrix extends AbstractTriangBandMatrix {
+
+ /**
+ * Constructor for UpperTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ */
+ public UpperTriangBandMatrix(int n, int kd) {
+ super(n, 0, kd, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ */
+ public UpperTriangBandMatrix(Matrix A, int kd) {
+ this(A, kd, true);
+ }
+
+ /**
+ * Constructor for UpperTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ public UpperTriangBandMatrix(Matrix A, int kd, boolean deep) {
+ super(A, 0, kd, deep, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangBandMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ */
+ UpperTriangBandMatrix(int n, int kd, Diag diag) {
+ super(n, 0, kd, UpLo.Upper, diag);
+ }
+
+ /**
+ * Constructor for UpperTriangBandMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the parts of <code>A</code>
+ * that lie within the allocated band are copied over, the rest
+ * is ignored
+ * @param kd
+ * Number of bands above the main diagonal (superdiagonals)
+ * @param deep
+ * True for a deep copy. For shallow copies, <code>A</code>
+ * must be a banded matrix
+ */
+ UpperTriangBandMatrix(Matrix A, int kd, boolean deep, Diag diag) {
+ super(A, 0, kd, deep, UpLo.Upper, diag);
+ }
+
+ @Override
+ public UpperTriangBandMatrix copy() {
+ return new UpperTriangBandMatrix(this, ku);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperTriangDenseMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperTriangDenseMatrix.java
new file mode 100644
index 0000000..c983648
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperTriangDenseMatrix.java
@@ -0,0 +1,175 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper triangular dense matrix. It has the same storage layout as the
+ * {@link no.uib.cipr.matrix.DenseMatrix DenseMatrix}, but only refers to
+ * elements above or on the main diagonal. The remaining elements are assumed to
+ * be zero, but since they are never accessed, they need not be.
+ */
+public class UpperTriangDenseMatrix extends AbstractTriangDenseMatrix {
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperTriangDenseMatrix(int n) {
+ super(n, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ UpperTriangDenseMatrix(int n, Diag diag) {
+ super(n, UpLo.Upper, diag);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ */
+ public UpperTriangDenseMatrix(Matrix A) {
+ this(A, Math.min(A.numRows(), A.numColumns()));
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ * @param deep
+ * True for deep copy, false for reference (in which case
+ * <code>A</code> must be a dense matrix)
+ */
+ public UpperTriangDenseMatrix(Matrix A, boolean deep) {
+ this(A, Math.min(A.numRows(), A.numColumns()), deep);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ * @param deep
+ * True for deep copy, false for reference (in which case
+ * <code>A</code> must be a dense matrix)
+ */
+ UpperTriangDenseMatrix(Matrix A, boolean deep, Diag diag) {
+ this(A, Math.min(A.numRows(), A.numColumns()), deep, diag);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ */
+ public UpperTriangDenseMatrix(Matrix A, int k) {
+ this(A, k, true);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ * @param deep
+ * True for deep copy, false for reference (in which case
+ * <code>A</code> must be a dense matrix)
+ */
+ public UpperTriangDenseMatrix(Matrix A, int k, boolean deep) {
+ super(A, k, deep, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangDenseMatrix
+ *
+ * @param A
+ * Matrix to copy from. Only the upper triangular part is copied
+ * @param k
+ * Size of matrix to refer.
+ * <code>k<min(numRows,numColumns)</code>
+ * @param deep
+ * True for deep copy, false for reference (in which case
+ * <code>A</code> must be a dense matrix)
+ */
+ UpperTriangDenseMatrix(Matrix A, int k, boolean deep, Diag diag) {
+ super(A, k, deep, UpLo.Upper, diag);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row > column)
+ throw new IllegalArgumentException("row > column");
+ super.add(row, column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row > column)
+ return 0;
+ return super.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row > column)
+ throw new IllegalArgumentException("row > column");
+ super.set(row, column, value);
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() <= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public Matrix set(Matrix A) {
+ zero();
+ copy(A);
+ return this;
+ }
+
+ @Override
+ public UpperTriangDenseMatrix copy() {
+ return new UpperTriangDenseMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/UpperTriangPackMatrix.java b/src/main/java/no/uib/cipr/matrix/UpperTriangPackMatrix.java
new file mode 100644
index 0000000..e4b45f4
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/UpperTriangPackMatrix.java
@@ -0,0 +1,181 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+
+/**
+ * Upper triangular packed matrix. In contrast with
+ * {@link no.uib.cipr.matrix.LowerTriangDenseMatrix LowerTriangDenseMatrix},
+ * this matrix exploits the sparsity by only storing about half the matrix. As
+ * such, the triangular matrix
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>12</sub></td>
+ * <td>a<sub>13</sub></td>
+ * <td>a<sub>14</sub></td>
+ * </tr>
+ * <tr>
+ * <td> </td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td>a<sub>24</sub></td>
+ * </tr>
+ * <tr>
+ * <td> </td>
+ * <td> </td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>34</sub></td>
+ * </tr>
+ * <tr>
+ * <td> </td>
+ * <td> </td>
+ * <td> </td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ * <p>
+ * is packed as follows:
+ * </p>
+ * <p>
+ * <table border="1">
+ * <tr>
+ * <td>a<sub>11</sub></td>
+ * <td>a<sub>12</sub></td>
+ * <td>a<sub>22</sub></td>
+ * <td>a<sub>13</sub></td>
+ * <td>a<sub>23</sub></td>
+ * <td>a<sub>33</sub></td>
+ * <td>a<sub>14</sub></td>
+ * <td>a<sub>24</sub></td>
+ * <td>a<sub>34</sub></td>
+ * <td>a<sub>44</sub></td>
+ * </tr>
+ * </table>
+ * </p>
+ */
+public class UpperTriangPackMatrix extends AbstractTriangPackMatrix {
+
+ /**
+ * Constructor for UpperTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ public UpperTriangPackMatrix(int n) {
+ super(n, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangPackMatrix
+ *
+ * @param n
+ * Size of the matrix. Since the matrix must be square, this
+ * equals both the number of rows and columns
+ */
+ UpperTriangPackMatrix(int n, Diag diag) {
+ super(n, UpLo.Upper, diag);
+ }
+
+ /**
+ * Constructor for UpperTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ */
+ public UpperTriangPackMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Constructor for UpperTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ public UpperTriangPackMatrix(Matrix A, boolean deep) {
+ super(A, deep, UpLo.Upper, Diag.NonUnit);
+ }
+
+ /**
+ * Constructor for UpperTriangPackMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. Only the entries of the relevant
+ * part are copied
+ * @param deep
+ * True if the copy is deep, else false (giving a shallow copy).
+ * For shallow copies, <code>A</code> must be a packed matrix
+ */
+ UpperTriangPackMatrix(Matrix A, boolean deep, Diag diag) {
+ super(A, deep, UpLo.Upper, diag);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ if (row > column)
+ throw new IllegalArgumentException("row > column");
+ data[getIndex(row, column)] += value;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ if (row > column)
+ throw new IllegalArgumentException("row > column");
+ data[getIndex(row, column)] = value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ if (row > column)
+ return 0;
+ return data[getIndex(row, column)];
+ }
+
+ /**
+ * Checks the row and column indices, and returns the linear data index
+ */
+ int getIndex(int row, int column) {
+ check(row, column);
+ return row + (column + 1) * column / 2;
+ }
+
+ @Override
+ void copy(Matrix A) {
+ for (MatrixEntry e : A)
+ if (e.row() <= e.column())
+ set(e.row(), e.column(), e.get());
+ }
+
+ @Override
+ public UpperTriangPackMatrix copy() {
+ return new UpperTriangPackMatrix(this);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/Vector.java b/src/main/java/no/uib/cipr/matrix/Vector.java
new file mode 100644
index 0000000..337536c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/Vector.java
@@ -0,0 +1,185 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.io.Serializable;
+
+/**
+ * Basic vector interface. It holds <code>double</code>s in an array, and is
+ * used alongside <code>Matrix</code> in numerical computations. Implementing
+ * classes decides on the actual storage.
+ *
+ * <h4>Basic operations</h4>
+ * <p>
+ * Use <code>size</code> to get the vector size. <code>get(int)</code> gets
+ * an element, and there are corresponding <code>set(int,double)</code> and
+ * <code>add(int,double)</code> methods as well. Note that vector indices are
+ * zero-based (typical for Java and C). This means that they range from 0 to
+ * <code>size-1</code>. It is legal to have <code>size</code> equal zero.
+ * </p>
+ * <p>
+ * Other basic operations are <code>zero</code> which zeros all the entries of
+ * the vector, which can be cheaper than either zeroing the vector manually, or
+ * creating a new vector, and the operation <code>copy</code> which creates a
+ * deep copy of the vector. This copy has separate storage, but starts with the
+ * same contents as the current vector.
+ * </p>
+ * <h4>Iterators</h4>
+ * <p>
+ * The vector interface extends <code>Iterable</code>, and the iterator
+ * returns a <code>VectorEntry</code> which contains current index and entry
+ * value. Note that the iterator may skip non-zero entries. Using an iterator,
+ * many simple and efficient algorithms can be created. The iterator also
+ * permits changing values in the vector, however only non-zero entries can be
+ * changed.
+ * </p>
+ * <h4>Basic linear algebra</h4>
+ * <p>
+ * A selection of basic linear algebra operations are available. To ensure high
+ * efficiency, little or no internal memory allocation is done, and the user is
+ * required to supply the output arguments.
+ * </p>
+ * <p>
+ * The operations available include:
+ * </p>
+ * <dl>
+ * <dt><i>Additions</i></dt>
+ * <dd>Vectors can be added to each other, even if their underlying vector
+ * structures are incompatible</dd>
+ * <dt><i>Scaling</i></dt>
+ * <dd>Scalar multiplication (scaling) of a whole vector</dd>
+ * <dt><i>Norms</i></dt>
+ * <dd>Both innerproducts and norms can be computed. Several common norms are
+ * supported</dd>
+ * </dl>
+ */
+public interface Vector extends Iterable<VectorEntry>, Serializable {
+
+ /**
+ * Size of the vector
+ */
+ int size();
+
+ /**
+ * <code>x(index) = value</code>
+ */
+ void set(int index, double value);
+
+ /**
+ * <code>x(index) += value</code>
+ */
+ void add(int index, double value);
+
+ /**
+ * Returns <code>x(index)</code>
+ */
+ double get(int index);
+
+ /**
+ * Creates a deep copy of the vector
+ */
+ Vector copy();
+
+ /**
+ * Zeros all the entries in the vector, while preserving any underlying
+ * structure
+ */
+ Vector zero();
+
+ /**
+ * <code>x=alpha*x</code>
+ *
+ * @return x
+ */
+ Vector scale(double alpha);
+
+ /**
+ * <code>x=y</code>
+ *
+ * @return x
+ */
+ Vector set(Vector y);
+
+ /**
+ * <code>x=alpha*y</code>
+ *
+ * @return x
+ */
+ Vector set(double alpha, Vector y);
+
+ /**
+ * <code>x = y + x</code>
+ *
+ * @return x
+ */
+ Vector add(Vector y);
+
+ /**
+ * <code>x = alpha*y + x</code>
+ *
+ * @return x
+ */
+ Vector add(double alpha, Vector y);
+
+ /**
+ * <code>x<sup>T</sup>*y</code>
+ */
+ double dot(Vector y);
+
+ /**
+ * Computes the given norm of the vector
+ *
+ * @param type
+ * The type of norm to compute
+ */
+ double norm(Norm type);
+
+ /**
+ * Supported vector-norms. The difference between the two 2-norms is that
+ * one is fast, but can overflow, while the robust version is overflow
+ * resistant, but slower.
+ */
+ enum Norm {
+
+ /**
+ * Sum of the absolute values of the entries
+ */
+ One,
+
+ /**
+ * The root of sum of squares
+ */
+ Two,
+
+ /**
+ * As the 2 norm may overflow, an overflow resistant version is also
+ * available. Note that it may be slower.
+ */
+ TwoRobust,
+
+ /**
+ * Largest entry in absolute value
+ */
+ Infinity
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/VectorEntry.java b/src/main/java/no/uib/cipr/matrix/VectorEntry.java
new file mode 100644
index 0000000..c34ffdd
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/VectorEntry.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * An entry of a vector. Returned by the iterators over a vector structure
+ */
+public interface VectorEntry {
+
+ /**
+ * Returns the current index
+ */
+ int index();
+
+ /**
+ * Returns the value at the current index
+ */
+ double get();
+
+ /**
+ * Sets the value at the current index
+ */
+ void set(double value);
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/MatrixInfo.java b/src/main/java/no/uib/cipr/matrix/io/MatrixInfo.java
new file mode 100644
index 0000000..a1b21c6
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/MatrixInfo.java
@@ -0,0 +1,276 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+/**
+ * Contains information on a matrix in the <a
+ * href="http://math.nist.gov/MatrixMarket">Matrix Market</a> exchange format.
+ * Supports all valid matrices.
+ */
+public class MatrixInfo {
+
+ /**
+ * What kind of numbers are stored
+ */
+ public enum MatrixField {
+
+ /**
+ * Real numbers
+ */
+ Real,
+
+ /**
+ * Integers
+ */
+ Integer,
+
+ /**
+ * Complex numbers
+ */
+ Complex,
+
+ /**
+ * Pattern matrix. No numbers stored
+ */
+ Pattern;
+ }
+
+ /**
+ * Symmetry structure of the matrix, if any
+ */
+ public enum MatrixSymmetry {
+
+ /**
+ * General matrix, no symmetry
+ */
+ General,
+
+ /**
+ * Symmetrical matrix
+ */
+ Symmetric,
+
+ /**
+ * Skew symmetrical matrix
+ */
+ SkewSymmetric,
+
+ /**
+ * Hermitian matrix. Only applicable for complex entris
+ */
+ Hermitian;
+ }
+
+ /**
+ * True if the matrix is sparse, else false
+ */
+ private boolean sparse;
+
+ /**
+ * Type of data stored
+ */
+ private MatrixField field;
+
+ /**
+ * Matrix symmetry
+ */
+ private MatrixSymmetry symmetry;
+
+ /**
+ * Creates a specific type
+ *
+ * @param sparse
+ * True for sparse matrices, else false
+ * @param field
+ * Type of data stored
+ * @param symmetry
+ * Matrix symmetry
+ */
+ public MatrixInfo(boolean sparse, MatrixField field, MatrixSymmetry symmetry) {
+ this.sparse = sparse;
+ this.field = field;
+ this.symmetry = symmetry;
+
+ validate();
+ }
+
+ /**
+ * Validates the representation
+ */
+ private void validate() {
+ if (isDense() && isPattern())
+ throw new IllegalArgumentException(
+ "Matrix cannot be dense with pattern storage");
+ if (isReal() && isHermitian())
+ throw new IllegalArgumentException(
+ "Data cannot be real with hermitian symmetry");
+ if (!isComplex() && isHermitian())
+ throw new IllegalArgumentException(
+ "Data must be complex with hermitian symmetry");
+ if (isPattern() && isSkewSymmetric())
+ throw new IllegalArgumentException(
+ "Storage cannot be pattern and skew symmetrical");
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is in coordinate format, else
+ * <code>false</code>
+ */
+ public boolean isSparse() {
+ return sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is in coordinate format, else
+ * <code>false</code>
+ */
+ public boolean isCoordinate() {
+ return sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is in array format, else
+ * <code>false</code>
+ */
+ public boolean isDense() {
+ return !sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is in array format, else
+ * <code>false</code>
+ */
+ public boolean isArray() {
+ return !sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix stores real numbers, else
+ * <code>false</code>
+ */
+ public boolean isReal() {
+ return field == MatrixField.Real;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix stores integers, else
+ * <code>false</code>
+ */
+ public boolean isInteger() {
+ return field == MatrixField.Integer;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix stores complex numbers, else
+ * <code>false</code>
+ */
+ public boolean isComplex() {
+ return field == MatrixField.Complex;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix does not store any numbers,
+ * else <code>false</code>
+ */
+ public boolean isPattern() {
+ return field == MatrixField.Pattern;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix form is general, else
+ * <code>false</code>
+ */
+ public boolean isGeneral() {
+ return symmetry == MatrixSymmetry.General;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is symmetrical, else
+ * <code>false</code>
+ */
+ public boolean isSymmetric() {
+ return symmetry == MatrixSymmetry.Symmetric;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is skew-symmetrical, else
+ * <code>false</code>
+ */
+ public boolean isSkewSymmetric() {
+ return symmetry == MatrixSymmetry.SkewSymmetric;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is Hermitian, else
+ * <code>false</code>
+ */
+ public boolean isHermitian() {
+ return symmetry == MatrixSymmetry.Hermitian;
+ }
+
+ /**
+ * Returns a string representation of the specifier. Can be used to provide
+ * a header for writing to a file. It is a two-line output, which can look
+ * like this:
+ *
+ * <pre>
+ * %%MatrixMarket matrix coordinate real general
+ * </pre>
+ */
+ @Override
+ public String toString() {
+ StringBuilder buf = new StringBuilder();
+
+ buf.append("%%MatrixMarket matrix ");
+
+ if (isSparse())
+ buf.append("coordinate ");
+ else
+ buf.append("array ");
+
+ if (isReal())
+ buf.append("real ");
+ else if (isComplex())
+ buf.append("complex ");
+ else if (isPattern())
+ buf.append("pattern ");
+ else if (isInteger())
+ buf.append("integer ");
+ else
+ // This should never happen
+ throw new IllegalArgumentException("Unknown field specification");
+
+ if (isGeneral())
+ buf.append("general\n");
+ else if (isSymmetric())
+ buf.append("symmetric\n");
+ else if (isSkewSymmetric())
+ buf.append("skew-symmetric\n");
+ else if (isHermitian())
+ buf.append("Hermitian\n");
+ else
+ // This should never happen
+ throw new IllegalArgumentException("Unknown symmetry specification");
+
+ return buf.toString();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/MatrixSize.java b/src/main/java/no/uib/cipr/matrix/io/MatrixSize.java
new file mode 100644
index 0000000..1c2594a
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/MatrixSize.java
@@ -0,0 +1,120 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+/**
+ * Contains the size of a matrix stored in the <a
+ * href="http://math.nist.gov/MatrixMarket">Matrix Market</a> exchange format
+ */
+public class MatrixSize {
+
+ /**
+ * Number of rows
+ */
+ private int numRows;
+
+ /**
+ * Number of columns
+ */
+ private int numColumns;
+
+ /**
+ * Number of entries stored
+ */
+ private int numEntries;
+
+ /**
+ * Constructor for MatrixSize
+ *
+ * @param numRows
+ * Number of rows in the matrix
+ * @param numColumns
+ * Number of columns in the matrix
+ * @param info
+ * Info on the matrix
+ */
+ public MatrixSize(int numRows, int numColumns, MatrixInfo info) {
+ this.numRows = numRows;
+ this.numColumns = numColumns;
+
+ if (!info.isDense())
+ throw new IllegalArgumentException("Matrix must be dense");
+
+ if (info.isGeneral())
+ numEntries = numRows * numColumns;
+ else if (info.isSymmetric() || info.isHermitian())
+ numEntries = ((numRows * numColumns - numRows) / 2 + numRows);
+ else if (info.isSkewSymmetric())
+ numEntries = (numRows * numColumns - numRows) / 2;
+ }
+
+ /**
+ * Constructor for MatrixSize
+ *
+ * @param numRows
+ * Number of rows in the matrix
+ * @param numColumns
+ * Number of columns in the matrix
+ * @param numEntries
+ * Number of entries stored
+ */
+ public MatrixSize(int numRows, int numColumns, int numEntries) {
+ this.numRows = numRows;
+ this.numColumns = numColumns;
+ this.numEntries = numEntries;
+
+ // We do this to avoid overflows
+ long maxR = numRows, maxC = numColumns, max = maxR * maxC;
+ if (numEntries > max)
+ throw new IllegalArgumentException(
+ "numEntries > numRows*numColumns");
+ }
+
+ /**
+ * Returns the number of rows in the matrix
+ */
+ public int numRows() {
+ return numRows;
+ }
+
+ /**
+ * Returns the number of columns in the matrix
+ */
+ public int numColumns() {
+ return numColumns;
+ }
+
+ /**
+ * Returns the number of entries stored
+ */
+ public int numEntries() {
+ return numEntries;
+ }
+
+ /**
+ * Returns <code>true</code> if the matrix is square, else
+ * <code>false</code>
+ */
+ public boolean isSquare() {
+ return numRows == numColumns;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/MatrixVectorReader.java b/src/main/java/no/uib/cipr/matrix/io/MatrixVectorReader.java
new file mode 100644
index 0000000..dae1952
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/MatrixVectorReader.java
@@ -0,0 +1,658 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+import java.io.BufferedReader;
+import java.io.EOFException;
+import java.io.IOException;
+import java.io.Reader;
+import java.io.StreamTokenizer;
+import java.util.LinkedList;
+import java.util.List;
+
+/**
+ * Reads matrices and vectors
+ */
+public class MatrixVectorReader extends BufferedReader {
+
+ /**
+ * Reads the entries of the matrix or vector
+ */
+ private StreamTokenizer st;
+
+ /**
+ * Constructor for MatrixVectorReader
+ *
+ * @param in
+ * A Reader
+ */
+ public MatrixVectorReader(Reader in) {
+ super(in);
+ setup();
+ }
+
+ /**
+ * Constructor for MatrixVectorReader
+ *
+ * @param in
+ * A Reader
+ * @param sz
+ * Input buffer size
+ */
+ public MatrixVectorReader(Reader in, int sz) {
+ super(in, sz);
+ setup();
+ }
+
+ /**
+ * Sets up the stream tokenizer
+ */
+ private void setup() {
+ st = new StreamTokenizer(this);
+ st.resetSyntax();
+ st.eolIsSignificant(false);
+ st.lowerCaseMode(true);
+
+ // Parse numbers as words
+ st.wordChars('0', '9');
+ st.wordChars('-', '.');
+
+ // Characters as words
+ st.wordChars('\u0000', '\u00FF');
+
+ // Skip comments
+ st.commentChar('%');
+
+ // Skip whitespace and newlines
+ st.whitespaceChars(' ', ' ');
+ st.whitespaceChars('\u0009', '\u000e');
+ }
+
+ /**
+ * Shifts the indices. Useful for converting between 0- and 1-based
+ * indicing.
+ *
+ * @param num
+ * Added to every index
+ * @param indices
+ * Indices to shift
+ */
+ public void add(int num, int[] indices) {
+ for (int i = 0; i < indices.length; ++i)
+ indices[i] += num;
+ }
+
+ /**
+ * Reads a line, and trims it of surrounding whitespace
+ *
+ * @throws IOException
+ * If either I/O errors occur, or there was nothing to read
+ */
+ private String readTrimmedLine() throws IOException {
+ String line = readLine();
+ if (line != null)
+ return line.trim();
+ else
+ throw new EOFException();
+ }
+
+ /**
+ * Reads the matrix info for the Matrix Market exchange format. The line
+ * must consist of exactly 5 space-separated entries, the first being
+ * "%%MatrixMarket"
+ */
+ public MatrixInfo readMatrixInfo() throws IOException {
+ String[] component = readTrimmedLine().split(" +");
+ if (component.length != 5)
+ throw new IOException(
+ "Current line unparsable. It must consist of 5 tokens");
+
+ // Read header
+ if (!component[0].equalsIgnoreCase("%%MatrixMarket"))
+ throw new IOException("Not in Matrix Market exchange format");
+
+ // This will always be "matrix"
+ if (!component[1].equalsIgnoreCase("matrix"))
+ throw new IOException("Expected \"matrix\", got " + component[1]);
+
+ // Sparse or dense?
+ boolean sparse = false;
+ if (component[2].equalsIgnoreCase("coordinate"))
+ sparse = true;
+ else if (component[2].equalsIgnoreCase("array"))
+ sparse = false;
+ else
+ throw new IOException("Unknown layout " + component[2]);
+
+ // Dataformat
+ MatrixInfo.MatrixField field = null;
+ if (component[3].equalsIgnoreCase("real"))
+ field = MatrixInfo.MatrixField.Real;
+ else if (component[3].equalsIgnoreCase("integer"))
+ field = MatrixInfo.MatrixField.Integer;
+ else if (component[3].equalsIgnoreCase("complex"))
+ field = MatrixInfo.MatrixField.Complex;
+ else if (component[3].equalsIgnoreCase("pattern"))
+ field = MatrixInfo.MatrixField.Pattern;
+ else
+ throw new IOException("Unknown field specification " + component[3]);
+
+ // Matrix pattern
+ MatrixInfo.MatrixSymmetry symmetry = null;
+ if (component[4].equalsIgnoreCase("general"))
+ symmetry = MatrixInfo.MatrixSymmetry.General;
+ else if (component[4].equalsIgnoreCase("symmetric"))
+ symmetry = MatrixInfo.MatrixSymmetry.Symmetric;
+ else if (component[4].equalsIgnoreCase("skew-symmetric"))
+ symmetry = MatrixInfo.MatrixSymmetry.SkewSymmetric;
+ else if (component[4].equalsIgnoreCase("Hermitian"))
+ symmetry = MatrixInfo.MatrixSymmetry.Hermitian;
+ else
+ throw new IOException("Unknown symmetry specification "
+ + component[4]);
+
+ // Pack together. This also verifies the format
+ return new MatrixInfo(sparse, field, symmetry);
+ }
+
+ /**
+ * Reads the vector info for the Matrix Market exchange format. The line
+ * must consist of exactly 4 space-separated entries, the first being
+ * "%%MatrixMarket"
+ */
+ public VectorInfo readVectorInfo() throws IOException {
+ String[] component = readTrimmedLine().split(" +");
+ if (component.length != 4)
+ throw new IOException(
+ "Current line unparsable. It must consist of 4 tokens");
+
+ // Read header
+ if (!component[0].equalsIgnoreCase("%%MatrixMarket"))
+ throw new IOException("Not in Matrix Market exchange format");
+
+ // This will always be "vector"
+ if (!component[1].equalsIgnoreCase("vector"))
+ throw new IOException("Expected \"vector\", got " + component[1]);
+
+ // Sparse or dense?
+ boolean sparse = false;
+ if (component[2].equalsIgnoreCase("coordinate"))
+ sparse = true;
+ else if (component[2].equalsIgnoreCase("array"))
+ sparse = false;
+ else
+ throw new IOException("Unknown layout " + component[2]);
+
+ // Dataformat
+ VectorInfo.VectorField field = null;
+ if (component[3].equalsIgnoreCase("real"))
+ field = VectorInfo.VectorField.Real;
+ else if (component[3].equalsIgnoreCase("integer"))
+ field = VectorInfo.VectorField.Integer;
+ else if (component[3].equalsIgnoreCase("complex"))
+ field = VectorInfo.VectorField.Complex;
+ else if (component[3].equalsIgnoreCase("pattern"))
+ field = VectorInfo.VectorField.Pattern;
+ else
+ throw new IOException("Unknown field specification " + component[3]);
+
+ // Pack together. This also verifies the format
+ return new VectorInfo(sparse, field);
+ }
+
+ /**
+ * Checks if a Matrix Market header is present ("%%MatrixMarket")
+ *
+ * @return True if a header was found, else false
+ * @throws IOException
+ */
+ public boolean hasInfo() throws IOException {
+ // Read a line, then skip back
+ mark(1024);
+ String[] component = readTrimmedLine().split(" +");
+ reset();
+
+ return component[0].equalsIgnoreCase("%%MatrixMarket");
+ }
+
+ /**
+ * Reads all the comments (lines starting with '%'). Positions the reader at
+ * the first non-comment line. Can only be called after reading the matrix
+ * or vector info. The comments read does not include '%' or the newline
+ */
+ public String[] readComments() throws IOException {
+ List<String> list = new LinkedList<String>();
+ while (true) {
+ mark(1024); // Line length equal 1024 at most
+ String line = readTrimmedLine();
+ if (line.length() > 0)
+ if (line.charAt(0) != '%') {
+ reset();
+ break;
+ } else
+ list.add(line.substring(1));
+ }
+ return list.toArray(new String[list.size()]);
+ }
+
+ /**
+ * Reads in the size of a matrix. Skips initial comments
+ */
+ public MatrixSize readMatrixSize(MatrixInfo info) throws IOException {
+ // Always read the matrix size
+ int numRows = getInt(), numColumns = getInt();
+ int size = getInt();
+
+ // For coordinate matrices we also read the number of entries
+ if (info.isDense())
+ return new MatrixSize(numRows, numColumns, info);
+ else {
+ int numEntries = getInt();
+ return new MatrixSize(numRows, numColumns, numEntries);
+ }
+ }
+
+ /**
+ * Reads in the size of an array matrix. Skips initial comments
+ */
+ public MatrixSize readArraySize() throws IOException {
+ int numRows = getInt(), numColumns = getInt();
+
+ return new MatrixSize(numRows, numColumns, numRows * numColumns);
+ }
+
+ /**
+ * Reads in the size of a coordinate matrix. Skips initial comments
+ */
+ public MatrixSize readCoordinateSize() throws IOException {
+ int numRows = getInt(), numColumns = getInt(), numEntries = getInt();
+
+ return new MatrixSize(numRows, numColumns, numEntries);
+ }
+
+ /**
+ * Reads in the size of a vector. Skips initial comments
+ */
+ public VectorSize readVectorSize(VectorInfo info) throws IOException {
+ // Always read the vector size
+ int size = getInt();
+
+ // For coordinate vectors we also read the number of entries
+ if (info.isDense())
+ return new VectorSize(size);
+ else {
+ int numEntries = getInt();
+ return new VectorSize(size, numEntries);
+ }
+ }
+
+ /**
+ * Reads in the size of a dense vector. Skips initial comments
+ */
+ public VectorSize readVectorArraySize() throws IOException {
+ int size = getInt();
+
+ return new VectorSize(size);
+ }
+
+ /**
+ * Reads in the size of a coordinate vector. Skips initial comments
+ */
+ public VectorSize readVectorCoordinateSize() throws IOException {
+ int size = getInt(), numEntries = getInt();
+
+ return new VectorSize(size, numEntries);
+ }
+
+ /**
+ * Reads the array data
+ */
+ public void readArray(double[] data) throws IOException {
+ int size = data.length;
+ for (int i = 0; i < size; ++i)
+ data[i] = getDouble();
+ }
+
+ /**
+ * Reads the array data
+ */
+ public void readArray(float[] data) throws IOException {
+ int size = data.length;
+ for (int i = 0; i < size; ++i)
+ data[i] = getFloat();
+ }
+
+ /**
+ * Reads the array data
+ */
+ public void readArray(int[] data) throws IOException {
+ int size = data.length;
+ for (int i = 0; i < size; ++i)
+ data[i] = getInt();
+ }
+
+ /**
+ * Reads the array data
+ */
+ public void readArray(long[] data) throws IOException {
+ int size = data.length;
+ for (int i = 0; i < size; ++i)
+ data[i] = getLong();
+ }
+
+ /**
+ * Reads the array data. The first array will contain real entries, while
+ * the second contain imaginary entries
+ */
+ public void readArray(double[] dataR, double[] dataI) throws IOException {
+ int size = dataR.length;
+ if (size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ dataR[i] = getDouble();
+ dataI[i] = getDouble();
+ }
+ }
+
+ /**
+ * Reads the array data. The first array will contain real entries, while
+ * the second contain imaginary entries
+ */
+ public void readArray(float[] dataR, float[] dataI) throws IOException {
+ int size = dataR.length;
+ if (size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ dataR[i] = getFloat();
+ dataI[i] = getFloat();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector
+ */
+ public void readCoordinate(int[] index, double[] data) throws IOException {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ data[i] = getDouble();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector
+ */
+ public void readCoordinate(int[] index, float[] data) throws IOException {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ data[i] = getFloat();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector
+ */
+ public void readCoordinate(int[] index, int[] data) throws IOException {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ data[i] = getInt();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector
+ */
+ public void readCoordinate(int[] index, long[] data) throws IOException {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ data[i] = getLong();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector. First data array contains real entries, and
+ * the second contains imaginary entries
+ */
+ public void readCoordinate(int[] index, float[] dataR, float[] dataI)
+ throws IOException {
+ int size = index.length;
+ if (size != dataR.length || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ dataR[i] = getFloat();
+ dataI[i] = getFloat();
+ }
+ }
+
+ /**
+ * Reads a coordinate vector. First data array contains real entries, and
+ * the second contains imaginary entries
+ */
+ public void readCoordinate(int[] index, double[] dataR, double[] dataI)
+ throws IOException {
+ int size = index.length;
+ if (size != dataR.length || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ index[i] = getInt();
+ dataR[i] = getDouble();
+ dataI[i] = getDouble();
+ }
+ }
+
+ /**
+ * Reads a pattern vector
+ */
+ public void readPattern(int[] index) throws IOException {
+ int size = index.length;
+ for (int i = 0; i < size; ++i)
+ index[i] = getInt();
+ }
+
+ /**
+ * Reads a coordinate matrix
+ */
+ public void readCoordinate(int[] row, int[] column, double[] data)
+ throws IOException {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ data[i] = getDouble();
+ }
+ }
+
+ /**
+ * Reads a coordinate matrix
+ */
+ public void readCoordinate(int[] row, int[] column, float[] data)
+ throws IOException {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ data[i] = getFloat();
+ }
+ }
+
+ /**
+ * Reads a coordinate matrix
+ */
+ public void readCoordinate(int[] row, int[] column, int[] data)
+ throws IOException {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ data[i] = getInt();
+ }
+ }
+
+ /**
+ * Reads a coordinate matrix
+ */
+ public void readCoordinate(int[] row, int[] column, long[] data)
+ throws IOException {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ data[i] = getLong();
+ }
+ }
+
+ /**
+ * Reads a pattern matrix
+ */
+ public void readPattern(int[] row, int[] column) throws IOException {
+ int size = row.length;
+ if (size != column.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ }
+ }
+
+ /**
+ * Reads a coordinate matrix. First data array contains real entries, and
+ * the second contains imaginary entries
+ */
+ public void readCoordinate(int[] row, int[] column, double[] dataR,
+ double[] dataI) throws IOException {
+ int size = row.length;
+ if (size != column.length || size != dataR.length
+ || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ dataR[i] = getDouble();
+ dataI[i] = getDouble();
+ }
+ }
+
+ /**
+ * Reads a coordinate matrix. First data array contains real entries, and
+ * the second contains imaginary entries
+ */
+ public void readCoordinate(int[] row, int[] column, float[] dataR,
+ float[] dataI) throws IOException {
+ int size = row.length;
+ if (size != column.length || size != dataR.length
+ || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i) {
+ row[i] = getInt();
+ column[i] = getInt();
+ dataR[i] = getFloat();
+ dataI[i] = getFloat();
+ }
+ }
+
+ /**
+ * Reads an integer
+ */
+ private int getInt() throws IOException {
+ st.nextToken();
+ if (st.ttype == StreamTokenizer.TT_WORD)
+ return Double.valueOf(st.sval).intValue();
+ else if (st.ttype == StreamTokenizer.TT_EOF)
+ throw new EOFException("End-of-File encountered during parsing");
+ else
+ throw new IOException("Unknown token found during parsing");
+ }
+
+ /**
+ * Reads a long
+ */
+ private long getLong() throws IOException {
+ st.nextToken();
+ if (st.ttype == StreamTokenizer.TT_WORD)
+ return Long.parseLong(st.sval);
+ else if (st.ttype == StreamTokenizer.TT_EOF)
+ throw new EOFException("End-of-File encountered during parsing");
+ else
+ throw new IOException("Unknown token found during parsing");
+ }
+
+ /**
+ * Reads a double
+ */
+ private double getDouble() throws IOException {
+ st.nextToken();
+ if (st.ttype == StreamTokenizer.TT_WORD)
+ return Double.parseDouble(st.sval);
+ else if (st.ttype == StreamTokenizer.TT_EOF)
+ throw new EOFException("End-of-File encountered during parsing");
+ else
+ throw new IOException("Unknown token found during parsing");
+ }
+
+ /**
+ * Reads a float
+ */
+ private float getFloat() throws IOException {
+ st.nextToken();
+ if (st.ttype == StreamTokenizer.TT_WORD)
+ return Float.parseFloat(st.sval);
+ else if (st.ttype == StreamTokenizer.TT_EOF)
+ throw new EOFException("End-of-File encountered during parsing");
+ else
+ throw new IOException("Unknown token found during parsing");
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/MatrixVectorWriter.java b/src/main/java/no/uib/cipr/matrix/io/MatrixVectorWriter.java
new file mode 100644
index 0000000..9ad18d6
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/MatrixVectorWriter.java
@@ -0,0 +1,531 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+import java.io.OutputStream;
+import java.io.PrintWriter;
+import java.io.Writer;
+import java.util.Locale;
+
+/**
+ * Writes matrices and vectors
+ */
+public class MatrixVectorWriter extends PrintWriter {
+
+ /**
+ * Constructor for MatrixVectorWriter
+ *
+ * @param out
+ */
+ public MatrixVectorWriter(OutputStream out) {
+ super(out);
+ }
+
+ /**
+ * Constructor for MatrixVectorWriter
+ *
+ * @param out
+ * @param autoFlush
+ */
+ public MatrixVectorWriter(OutputStream out, boolean autoFlush) {
+ super(out, autoFlush);
+ }
+
+ /**
+ * Constructor for MatrixVectorWriter
+ *
+ * @param out
+ */
+ public MatrixVectorWriter(Writer out) {
+ super(out);
+ }
+
+ /**
+ * Constructor for MatrixVectorWriter
+ *
+ * @param out
+ * @param autoFlush
+ */
+ public MatrixVectorWriter(Writer out, boolean autoFlush) {
+ super(out, autoFlush);
+ }
+
+ /**
+ * Shifts the indices. Useful for converting between 0- and 1-based
+ * indicing.
+ *
+ * @param num
+ * Added to every index
+ * @param indices
+ * Indices to shift
+ */
+ public void add(int num, int[] indices) {
+ for (int i = 0; i < indices.length; ++i)
+ indices[i] += num;
+ }
+
+ /**
+ * Prints the matrix info
+ */
+ public void printMatrixInfo(MatrixInfo info) {
+ print(info.toString());
+ }
+
+ /**
+ * Prints the vector info
+ */
+ public void printVectorInfo(VectorInfo info) {
+ print(info.toString());
+ }
+
+ /**
+ * Prints the matrix size
+ */
+ public void printMatrixSize(MatrixSize size, MatrixInfo info) {
+ format(Locale.ENGLISH, "%10d %10d", size.numRows(), size.numColumns());
+ if (info.isCoordinate())
+ format(Locale.ENGLISH, " %19d", size.numEntries());
+ println();
+ }
+
+ /**
+ * Prints the matrix size. Assumes coordinate format
+ */
+ public void printMatrixSize(MatrixSize size) {
+ format(Locale.ENGLISH, "%10d %10d %19d\n", size.numRows(), size.numColumns(), size
+ .numEntries());
+ }
+
+ /**
+ * Prints the vector size
+ */
+ public void printVectorSize(VectorSize size, VectorInfo info) {
+ format(Locale.ENGLISH, "%10d", size.size());
+ if (info.isCoordinate())
+ format(Locale.ENGLISH, " %19d", size.numEntries());
+ println();
+ }
+
+ /**
+ * Prints the vector size. Assumes coordinate format
+ */
+ public void printVectorSize(VectorSize size) {
+ format(Locale.ENGLISH, "%10d %19d\n", size.size(), size.numEntries());
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line.
+ */
+ public void printArray(float[] data) {
+ for (int i = 0; i < data.length; ++i)
+ format(Locale.ENGLISH, "% .12e\n", data[i]);
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line.
+ */
+ public void printArray(double[] data) {
+ for (int i = 0; i < data.length; ++i)
+ format(Locale.ENGLISH, "% .12e\n", data[i]);
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line. The first
+ * array specifies the real entries, and the second is the imaginary entries
+ */
+ public void printArray(float[] dataR, float[] dataI) {
+ int size = dataR.length;
+ if (size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "% .12e % .12e\n", dataR[i], dataI[i]);
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line. The first
+ * array specifies the real entries, and the second is the imaginary entries
+ */
+ public void printArray(double[] dataR, double[] dataI) {
+ int size = dataR.length;
+ if (size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "% .12e % .12e\n", dataR[i], dataI[i]);
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line.
+ */
+ public void printArray(int[] data) {
+ for (int i = 0; i < data.length; ++i)
+ format(Locale.ENGLISH, "%10d\n", data[i]);
+ }
+
+ /**
+ * Prints an array to the underlying stream. One entry per line.
+ */
+ public void printArray(long[] data) {
+ for (int i = 0; i < data.length; ++i)
+ format(Locale.ENGLISH, "%10d\n", data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to the index, typically, this can
+ * transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] index, float[] data, int offset) {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d % .12e\n", index[i] + offset, data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to the index, typically, this can
+ * transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] index, double[] data, int offset) {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d % .12e\n", index[i] + offset, data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to the index, typically, this can
+ * transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] index, int[] data, int offset) {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d\n", index[i] + offset, data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to the index, typically, this can
+ * transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] index, long[] data, int offset) {
+ int size = index.length;
+ if (size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d\n", index[i] + offset, data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] row, int[] column, float[] data,
+ int offset) {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d % .12e\n", row[i] + offset, column[i] + offset,
+ data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] row, int[] column, double[] data,
+ int offset) {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d % .12e\n", row[i] + offset, column[i] + offset,
+ data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based. The first float array
+ * specifies the real entries, and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] index, float[] dataR, float[] dataI,
+ int offset) {
+ int size = index.length;
+ if (size != dataR.length || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d % .12e % .12e\n", index[i] + offset, dataR[i],
+ dataI[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based. The first double
+ * array specifies the real entries, and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] index, double[] dataR, double[] dataI,
+ int offset) {
+ int size = index.length;
+ if (size != dataR.length || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d % .12e % .12e\n", index[i] + offset, dataR[i],
+ dataI[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based. The first float array
+ * specifies the real entries, and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] row, int[] column, float[] dataR,
+ float[] dataI, int offset) {
+ int size = row.length;
+ if (size != column.length || size != dataR.length
+ || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d % .12e % .12e\n", row[i] + offset, column[i]
+ + offset, dataR[i], dataI[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based. The first double
+ * array specifies the real entries, and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] row, int[] column, double[] dataR,
+ double[] dataI, int offset) {
+ int size = row.length;
+ if (size != column.length || size != dataR.length
+ || size != dataI.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d % .12e % .12e\n", row[i] + offset, column[i]
+ + offset, dataR[i], dataI[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] row, int[] column, int[] data, int offset) {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d %19d\n", row[i] + offset, column[i] + offset,
+ data[i]);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The offset is added to each index, typically, this
+ * can transform from a 0-based indicing to a 1-based.
+ */
+ public void printCoordinate(int[] row, int[] column, long[] data, int offset) {
+ int size = row.length;
+ if (size != column.length || size != data.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d %19d\n", row[i] + offset, column[i] + offset,
+ data[i]);
+ }
+
+ /**
+ * Prints the coordinates to the underlying stream. One index pair on each
+ * line. The offset is added to each index, typically, this can transform
+ * from a 0-based indicing to a 1-based.
+ */
+ public void printPattern(int[] row, int[] column, int offset) {
+ int size = row.length;
+ if (size != column.length)
+ throw new IllegalArgumentException(
+ "All arrays must be of the same size");
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d %10d\n", row[i] + offset, column[i] + offset);
+ }
+
+ /**
+ * Prints the coordinates to the underlying stream. One index on each line.
+ * The offset is added to each index, typically, this can transform from a
+ * 0-based indicing to a 1-based.
+ */
+ public void printPattern(int[] index, int offset) {
+ int size = index.length;
+ for (int i = 0; i < size; ++i)
+ format(Locale.ENGLISH, "%10d\n", index[i] + offset);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line
+ */
+ public void printCoordinate(int[] row, int[] column, float[] data) {
+ printCoordinate(row, column, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line
+ */
+ public void printCoordinate(int[] row, int[] column, double[] data) {
+ printCoordinate(row, column, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The first double array specifies the real entries,
+ * and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] row, int[] column, float[] dataR,
+ float[] dataI) {
+ printCoordinate(row, column, dataR, dataI, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line. The first double array specifies the real entries,
+ * and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] row, int[] column, double[] dataR,
+ double[] dataI) {
+ printCoordinate(row, column, dataR, dataI, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line
+ */
+ public void printCoordinate(int[] row, int[] column, int[] data) {
+ printCoordinate(row, column, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index pair and
+ * entry on each line
+ */
+ public void printCoordinate(int[] row, int[] column, long[] data) {
+ printCoordinate(row, column, data, 0);
+ }
+
+ /**
+ * Prints the coordinates to the underlying stream. One index pair on each
+ * line
+ */
+ public void printPattern(int[] row, int[] column) {
+ printPattern(row, column, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line
+ */
+ public void printCoordinate(int[] index, float[] data) {
+ printCoordinate(index, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line
+ */
+ public void printCoordinate(int[] index, double[] data) {
+ printCoordinate(index, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The first double array specifies the real entries,
+ * and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] index, float[] dataR, float[] dataI) {
+ printCoordinate(index, dataR, dataI, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line. The first double array specifies the real entries,
+ * and the second is the imaginary entries
+ */
+ public void printCoordinate(int[] index, double[] dataR, double[] dataI) {
+ printCoordinate(index, dataR, dataI, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line
+ */
+ public void printCoordinate(int[] index, int[] data) {
+ printCoordinate(index, data, 0);
+ }
+
+ /**
+ * Prints the coordinate format to the underlying stream. One index and
+ * entry on each line
+ */
+ public void printCoordinate(int[] index, long[] data) {
+ printCoordinate(index, data, 0);
+ }
+
+ /**
+ * Prints the coordinates to the underlying stream. One index on each line
+ */
+ public void printPattern(int[] index) {
+ printPattern(index, 0);
+ }
+
+ /**
+ * Prints all the comments. Prepends a '%' and appends a newline to every
+ * comment
+ */
+ public void printComments(String[] comments) {
+ for (String comment : comments)
+ println("%" + comment);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/VectorInfo.java b/src/main/java/no/uib/cipr/matrix/io/VectorInfo.java
new file mode 100644
index 0000000..9264746
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/VectorInfo.java
@@ -0,0 +1,187 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+/**
+ * Contains information on a vector in a variant of the <a
+ * href="http://math.nist.gov/MatrixMarket">Matrix Market</a> exchange format
+ */
+public class VectorInfo {
+
+ /**
+ * What kind of numbers are stored
+ */
+ public enum VectorField {
+ /**
+ * Real numbers
+ */
+ Real,
+
+ /**
+ * Integers
+ */
+ Integer,
+
+ /**
+ * Complex numbers
+ */
+ Complex,
+
+ /**
+ * Pattern matrix. No numbers stored
+ */
+ Pattern;
+ }
+
+ /**
+ * True if the vector is sparse, else false
+ */
+ private boolean sparse;
+
+ /**
+ * Type of data stored
+ */
+ private VectorField field;
+
+ /**
+ * Creates a specific type
+ *
+ * @param sparse
+ * True for sparse vectors, else false
+ * @param field
+ * Type of data stored
+ */
+ public VectorInfo(boolean sparse, VectorField field) {
+ this.sparse = sparse;
+ this.field = field;
+
+ validate();
+ }
+
+ /**
+ * Validates the representation
+ */
+ private void validate() {
+ if (isDense() && isPattern())
+ throw new IllegalArgumentException(
+ "Vector cannot be dense with pattern storage");
+ }
+
+ /**
+ * Returns <code>true</code> if the vector is in coordinate format, else
+ * <code>false</code>
+ */
+ public boolean isSparse() {
+ return sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector is in coordinate format, else
+ * <code>false</code>
+ */
+ public boolean isCoordinate() {
+ return sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector is in array format, else
+ * <code>false</code>
+ */
+ public boolean isDense() {
+ return !sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector is in array format, else
+ * <code>false</code>
+ */
+ public boolean isArray() {
+ return !sparse;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector stores real numbers, else
+ * <code>false</code>
+ */
+ public boolean isReal() {
+ return field == VectorField.Real;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector stores integers, else
+ * <code>false</code>
+ */
+ public boolean isInteger() {
+ return field == VectorField.Integer;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector stores complex numbers, else
+ * <code>false</code>
+ */
+ public boolean isComplex() {
+ return field == VectorField.Complex;
+ }
+
+ /**
+ * Returns <code>true</code> if the vector does not store any numbers,
+ * else <code>false</code>
+ */
+ public boolean isPattern() {
+ return field == VectorField.Pattern;
+ }
+
+ /**
+ * Returns a string representation of the specifier. Can be used to provide
+ * a header for writing to a file. It is a two-line output, which can look
+ * like this:
+ *
+ * <pre>
+ * %%MatrixMarket vector coordinate real
+ * </pre>
+ */
+ @Override
+ public String toString() {
+ StringBuilder buf = new StringBuilder();
+
+ buf.append("%%MatrixMarket vector ");
+
+ if (isSparse())
+ buf.append("coordinate ");
+ else
+ buf.append("array ");
+
+ if (isReal())
+ buf.append("real\n");
+ else if (isComplex())
+ buf.append("complex\n");
+ else if (isPattern())
+ buf.append("pattern\n");
+ else if (isInteger())
+ buf.append("integer\n");
+ else
+ // This should never happen
+ throw new IllegalArgumentException("Unknown field specification");
+
+ return buf.toString();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/VectorSize.java b/src/main/java/no/uib/cipr/matrix/io/VectorSize.java
new file mode 100644
index 0000000..440d4b1
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/VectorSize.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.io;
+
+/**
+ * Contains the size of a vectir stored in a variant of the <a
+ * href="http://math.nist.gov/MatrixMarket">Matrix Market</a> exchange format
+ */
+public class VectorSize {
+
+ /**
+ * Size of the vector
+ */
+ private int size;
+
+ /**
+ * Number of entries stored
+ */
+ private int numEntries;
+
+ /**
+ * Constructor for VectorSize. Assumes dense format
+ *
+ * @param size
+ * Size of the matrix
+ */
+ public VectorSize(int size) {
+ this.size = size;
+ numEntries = size;
+
+ if (size < 0)
+ throw new IllegalArgumentException("size < 0");
+ }
+
+ /**
+ * Constructor for VectorSize
+ *
+ * @param size
+ * Size of the matrix
+ * @param numEntries
+ * Number of entries stored
+ */
+ public VectorSize(int size, int numEntries) {
+ this.size = size;
+ this.numEntries = numEntries;
+
+ if (size < 0 || numEntries < 0)
+ throw new IllegalArgumentException("size < 0 || numEntries < 0");
+ if (numEntries > size)
+ throw new IllegalArgumentException("numEntries > size");
+ }
+
+ /**
+ * Returns the size of the vector
+ */
+ public int size() {
+ return size;
+ }
+
+ /**
+ * Returns the number of entries in the vector
+ */
+ public int numEntries() {
+ return numEntries;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/io/package.html b/src/main/java/no/uib/cipr/matrix/io/package.html
new file mode 100644
index 0000000..64b3dfc
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/io/package.html
@@ -0,0 +1,17 @@
+<html>
+<body>
+I/O operations with matrices and vectors. The formats supported are:
+<p>
+<ul>
+<li><a href="http://math.nist.gov/MatrixMarket/formats.html#coord">
+ Coordinate format</a></li>
+<li><a href="http://math.nist.gov/MatrixMarket/formats.html#MMformat">
+ Matrix Market exchange format</a></li>
+</ul>
+</p>
+<p>
+The <a href="http://math.nist.gov/MatrixMarket/formats.html#hb">Harwell-Boeing
+format</a> is not supported.
+</p>
+</body>
+</html>
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/overview.html b/src/main/java/no/uib/cipr/matrix/overview.html
new file mode 100644
index 0000000..4b64c05
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/overview.html
@@ -0,0 +1,11 @@
+<html>
+ <body>
+ <p>
+ A library for high performance scientific computing in Java.
+ </p>
+ <p>
+ Visit the <a href="http://rs.cipr.uib.no/mtj">homepage</a>
+ for the most recent releases, examples and documentation.
+ </p>
+ </body>
+</html>
diff --git a/src/main/java/no/uib/cipr/matrix/package.html b/src/main/java/no/uib/cipr/matrix/package.html
new file mode 100644
index 0000000..e7082d2
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/package.html
@@ -0,0 +1,131 @@
+<html>
+ <body>
+ Dense and structured sparse matrices, along with matrix factorisations
+ and solvers. The main components are:
+ <ul>
+ <li><strong>Matrix and vector interfaces</strong>
+ <ul>
+ <li><a href="Matrix.html">Matrix</a> -
+ Linear algebra matrix.</li>
+ <li><a href="Vector.html">Vector</a> -
+ Linear algebra vector.</li>
+ </ul>
+ </li>
+ <li><strong>Matrix and vector implementations</strong>
+ <ul>
+
+ <li><a href="DenseVector.html">DenseVector</a> -
+ Stores the vector in a dense array.</li>
+ </li>
+
+ <li><strong>Dense matrices</strong> - Stores the data columnwise in a linear array.
+ <ul>
+ <li><a href="DenseMatrix.html">DenseMatrix</a> -
+ Recommended matrix for general cases.</li>
+ <li><strong>{Lower,Upper}{Symm,SPD}DenseMatrix</strong> -
+ Assumes implicit symmetry, and that the matrix is positive definite (SPD).
+ Stores entries in either the upper or lower triangular part.</li>
+ <li><strong>[Unit]{Lower,Upper}TriangDenseMatrix</strong> -
+ Only stores in the lower or upper triangular part, and assumes the rest is zero.
+ If the matrix is unit, the main diagonal is implicitly assumes to be one.</li>
+ </ul></li>
+
+ <li><strong>Packed matrices</strong> - Stores either the lower or upper triangular part of the matrix.
+ <ul>
+ <li><strong>{Lower,Upper}{Symm,SPD}PackMatrix</strong> -
+ Like their dense counterparts, but consumes less memory.</li>
+ <li><strong>[Unit]{Lower,Upper}TriangPackMatrix</strong> -
+ Also like their dense counterparts, but consumes less memory.</li>
+ </ul></li>
+
+ <li><strong>Banded matrices</strong> - Stores the matrix in a dense band.
+ <ul>
+ <li><a href="BandMatrix.html">BandMatrix</a> -
+ General band matrix with the possibility of different upper and lower triangular bandwidths.</li>
+ <li><strong>{Lower,Upper}{Symm,SPD}BandMatrix</strong> -
+ Like their dense counterparts, but consumes less memory by only storing the lower or upper parts.</li>
+ <li><strong>[Unit]{Lower,Upper}TriangBandMatrix</strong> -
+ Also like their dense counterparts, but consumes less memory by only storing the lower or upper parts..</li>
+ </ul></li>
+
+ <li><strong>Tridiagonal matrices</strong> - Stores just three diagonals.
+ <ul>
+ <li><a href="TridiagMatrix.html">TridiagMatrix</a> -
+ Standard tridiagonal matrix.</li>
+ <li><strong>{Symm,SPD}TridiagMatrix</strong> -
+ Stores only the main diagonal and an off-diagonal.
+ May also assume that the matrix is positive definite.</li>
+ </ul></li>
+
+ </ul></li>
+
+ <li><strong>Abstract implementations</strong>
+ <ul>
+ <li><a href="AbstractMatrix.html">AbstractMatrix</a> -
+ Implements all the matrix operations, but doesn't specify the matrix storage.</li>
+ <li><a href="AbstractVector.html">AbstractVector</a> -
+ Implements all the vector operations, but doesn't specify the vector storage.</li>
+ </ul></li>
+
+ <li><strong>Utility methods and iterators</strong>
+ <ul>
+ <li><a href="Matrices.html">Matrices</a> -
+ Creates submatrices and synchronized matrices. Also has several
+ miscellaneous utility methods.</li>
+ <li><a href="MatrixEntry.html">MatrixEntry</a> -
+ Entry returned when iterating over a matrix using the for-each
+ statement.</li>
+ <li><a href="VectorEntry.html">VectorEntry</a> -
+ Entry returned when iterating over a vector using the for-each
+ statement.</li>
+ </ul></li>
+
+ <li><strong>LU decompositions</strong>
+ <ul>
+ <li><a href="BandLU.html">BandLU</a> -
+ LU decomposition of a banded matrix.</li>
+ <li><a href="DenseLU.html">DenseLU</a> -
+ LU decomposition of a dense matrix.</li>
+ <li><a href="BandCholesky.html">BandCholesky</a> -
+ Cholesky decomposition of a banded matrix.</li>
+ <li><a href="DenseCholesky.html">DenseCholesky</a> -
+ Cholesky decomposition of a dense SPD matrix.</li>
+ <li><a href="PackCholesky.html">PackCholesky</a> -
+ Cholesky decomposition of a packed SPD matrix.</li>
+ </ul>
+ </li>
+ <li><strong>Orthogonal decompositions</strong>
+ <ul>
+ <li><a href="LQ.html">LQ</a> -
+ LQ decomposition.</li>
+ <li><a href="QL.html">QL</a> -
+ QL decomposition.</li>
+ <li><a href="QR.html">QR</a> -
+ QR decomposition.</li>
+ <li><a href="RQ.html">RQ</a> -
+ RQ decomposition.</li>
+ <li><a href="GivensRotation.html">GivensRotation</a> -
+ Givens plane rotation.</li>
+ </ul>
+ </li>
+ <li><strong>Spectral decompositions</strong>
+ <ul>
+ <li><a href="EVD.html">EVD</a> -
+ Eigenvalue decomposition of general matrices.</li>
+ <li><a href="SVD.html">SVD</a> -
+ Singular value decomposition</li>
+ <li><a href="SymmBandEVD.html">SymmBandEVD</a> -
+ Eigenvalue decomposition of symmetrical, banded matrices.</li>
+ <li><a href="SymmDenseEVD.html">SymmDenseEVD</a> -
+ Eigenvalue decomposition of symmetrical, dense matrices.</li>
+ <li><a href="SymmPackEVD.html">SymmPackEVD</a> -
+ Eigenvalue decomposition of symmetrical, packed matrices.</li>
+ <li><a href="SymmTridiagEVD.html">SymmTridiagEVD</a> -
+ Eigenvalue decomposition of symmetrical, tridiagonal matrices.</li>
+ </ul>
+ </li>
+ </ul>
+ <p>
+ </p>
+ </body>
+</html>
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/AMG.java b/src/main/java/no/uib/cipr/matrix/sparse/AMG.java
new file mode 100644
index 0000000..f99c7c7
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/AMG.java
@@ -0,0 +1,929 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.ArrayList;
+import java.util.Arrays;
+import java.util.HashMap;
+import java.util.HashSet;
+import java.util.LinkedList;
+import java.util.List;
+import java.util.Map;
+import java.util.Set;
+
+import no.uib.cipr.matrix.DenseLU;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Algebraic multigrid preconditioner. Uses the smoothed aggregation method
+ * described by Vanek, Mandel, and Brezina (1996).
+ */
+public class AMG implements Preconditioner {
+
+ /**
+ * Relaxations at each level
+ */
+ private SSOR[] preM, postM;
+
+ /**
+ * The number of levels
+ */
+ private int m;
+
+ /**
+ * System matrix at each level, except at the coarsest
+ */
+ private CompRowMatrix[] A;
+
+ /**
+ * LU factorization at the coarsest level
+ */
+ private DenseLU lu;
+
+ /**
+ * Solution, right-hand side, and residual vectors at each level
+ */
+ private DenseVector[] u, f, r;
+
+ /**
+ * Interpolation operators going to a finer mesh
+ */
+ private CompColMatrix[] I;
+
+ /**
+ * Smallest matrix size before terminating the AMG setup phase. Matrices
+ * smaller than this will be solved by a direct solver
+ */
+ private final int min;
+
+ /**
+ * Number of times to perform the pre- and post-smoothings
+ */
+ private final int nu1, nu2;
+
+ /**
+ * Determines cycle type. gamma=1 is V, gamma=2 is W
+ */
+ private final int gamma;
+
+ /**
+ * Overrelaxation parameters in the pre- and post-smoothings, and with the
+ * possibility of distinct values in the forward and reverse sweeps
+ */
+ private final double omegaPreF, omegaPreR, omegaPostF, omegaPostR;
+
+ /**
+ * Perform a reverse (backwards) smoothing sweep
+ */
+ private final boolean reverse;
+
+ /**
+ * Jacobi damping parameter, between zero and one. If it equals zero, the
+ * method reduces to the standard aggregate multigrid method
+ */
+ private final double omega;
+
+ /**
+ * Operating in transpose mode?
+ */
+ private boolean transpose;
+
+ /**
+ * Sets up the algebraic multigrid preconditioner
+ *
+ * @param omegaPreF
+ * Overrelaxation parameter in the forward sweep of the
+ * pre-smoothing
+ * @param omegaPreR
+ * Overrelaxation parameter in the backwards sweep of the
+ * pre-smoothing
+ * @param omegaPostF
+ * Overrelaxation parameter in the forward sweep of the
+ * post-smoothing
+ * @param omegaPostR
+ * Overrelaxation parameter in the backwards sweep of the
+ * post-smoothing
+ * @param nu1
+ * Number of pre-relaxations to perform
+ * @param nu2
+ * Number of post-relaxations to perform
+ * @param gamma
+ * Number of times to go to a coarser level
+ * @param min
+ * Smallest matrix size before using a direct solver
+ * @param omega
+ * Jacobi damping parameter, between zero and one. If it equals
+ * zero, the method reduces to the standard aggregate multigrid
+ * method
+ */
+ public AMG(double omegaPreF, double omegaPreR, double omegaPostF,
+ double omegaPostR, int nu1, int nu2, int gamma, int min,
+ double omega) {
+ this.omegaPreF = omegaPreF;
+ this.omegaPreR = omegaPreR;
+ this.omegaPostF = omegaPostF;
+ this.omegaPostR = omegaPostR;
+
+ reverse = true;
+
+ this.nu1 = nu1;
+ this.nu2 = nu2;
+ this.gamma = gamma;
+ this.min = min;
+
+ this.omega = omega;
+ }
+
+ /**
+ * Sets up the algebraic multigrid preconditioner. Uses an SOR method,
+ * without the backward sweep in SSOR
+ *
+ * @param omegaPre
+ * Overrelaxation parameter in the pre-smoothing
+ * @param omegaPost
+ * Overrelaxation parameter in the post-smoothing
+ * @param nu1
+ * Number of pre-relaxations to perform
+ * @param nu2
+ * Number of post-relaxations to perform
+ * @param gamma
+ * Number of times to go to a coarser level
+ * @param min
+ * Smallest matrix size before using a direct solver
+ * @param omega
+ * Jacobi damping parameter, between zero and one. If it equals
+ * zero, the method reduces to the standard aggregate multigrid
+ * method
+ */
+ public AMG(double omegaPre, double omegaPost, int nu1, int nu2, int gamma,
+ int min, double omega) {
+ this.omegaPreF = omegaPre;
+ this.omegaPreR = omegaPre;
+ this.omegaPostF = omegaPost;
+ this.omegaPostR = omegaPost;
+
+ reverse = false;
+
+ this.nu1 = nu1;
+ this.nu2 = nu2;
+ this.gamma = gamma;
+ this.min = min;
+
+ this.omega = omega;
+ }
+
+ /**
+ * Sets up the algebraic multigrid preconditioner using some default
+ * parameters. In the presmoothing, <code>omegaF=1</code> and
+ * <code>omegaR=1.85</code>, while in the postsmoothing,
+ * <code>omegaF=1.85</code> and <code>omegaR=1</code>. Sets
+ * <code>nu1=nu2=gamma=1</code>, has a smallest matrix size of 40, and
+ * sets <code>omega=2/3</code>.
+ */
+ public AMG() {
+ this(1, 1.85, 1.85, 1, 1, 1, 1, 40, 2. / 3);
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ u[0].set(x);
+ f[0].set(b);
+
+ transpose = false;
+ cycle(0);
+
+ return x.set(u[0]);
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ u[0].set(x);
+ f[0].set(b);
+
+ transpose = true;
+ cycle(0);
+
+ return x.set(u[0]);
+ }
+
+ public void setMatrix(Matrix A) {
+ List<CompRowMatrix> Al = new LinkedList<CompRowMatrix>();
+ List<CompColMatrix> Il = new LinkedList<CompColMatrix>();
+
+ Al.add(new CompRowMatrix(A));
+
+ for (int k = 0; Al.get(k).numRows() > min; ++k) {
+
+ CompRowMatrix Af = Al.get(k);
+
+ double eps = 0.08 * Math.pow(0.5, k);
+
+ // Create the aggregates
+ Aggregator aggregator = new Aggregator(Af, eps);
+
+ // If no aggregates were created, no interpolation operator will be
+ // created, and the setup phase stops
+ if (aggregator.getAggregates().size() == 0)
+ break;
+
+ // Create an interpolation operator using smoothing. This also
+ // creates the Galerkin operator
+ Interpolator sa = new Interpolator(aggregator, Af, omega);
+
+ Al.add(sa.getGalerkinOperator());
+ Il.add(sa.getInterpolationOperator());
+ }
+
+ // Copy to array storage
+ m = Al.size();
+ if (m == 0)
+ throw new RuntimeException("Matrix too small for AMG");
+
+ I = new CompColMatrix[m - 1];
+ this.A = new CompRowMatrix[m - 1];
+
+ Il.toArray(I);
+ for (int i = 0; i < Al.size() - 1; ++i)
+ this.A[i] = Al.get(i);
+
+ // Create a LU decomposition of the smallest Galerkin matrix
+ DenseMatrix Ac = new DenseMatrix(Al.get(Al.size() - 1));
+ lu = new DenseLU(Ac.numRows(), Ac.numColumns());
+ lu.factor(Ac);
+
+ // Allocate vectors at each level
+ u = new DenseVector[m];
+ f = new DenseVector[m];
+ r = new DenseVector[m];
+ for (int k = 0; k < m; ++k) {
+ int n = Al.get(k).numRows();
+ u[k] = new DenseVector(n);
+ f[k] = new DenseVector(n);
+ r[k] = new DenseVector(n);
+ }
+
+ // Set up the SSOR relaxation schemes
+ preM = new SSOR[m - 1];
+ postM = new SSOR[m - 1];
+ for (int k = 0; k < m - 1; ++k) {
+ CompRowMatrix Ak = this.A[k];
+ preM[k] = new SSOR(Ak, reverse, omegaPreF, omegaPreR);
+ postM[k] = new SSOR(Ak, reverse, omegaPostF, omegaPostR);
+ preM[k].setMatrix(Ak);
+ postM[k].setMatrix(Ak);
+ }
+ }
+
+ /**
+ * Performs a multigrid cycle
+ *
+ * @param k
+ * Level to cycle at. Start by calling <code>cycle(0)</code>
+ */
+ private void cycle(int k) {
+ if (k == m - 1)
+ directSolve();
+ else {
+
+ // Presmoothings
+ preRelax(k);
+
+ u[k + 1].zero();
+
+ // Compute the residual
+ A[k].multAdd(-1, u[k], r[k].set(f[k]));
+
+ // Restrict to the next coarser level
+ I[k].transMult(r[k], f[k + 1]);
+
+ // Recurse to next level
+ for (int i = 0; i < gamma; ++i)
+ cycle(k + 1);
+
+ // Add residual correction by prolongation
+ I[k].multAdd(u[k + 1], u[k]);
+
+ // Postsmoothings
+ postRelax(k);
+ }
+ }
+
+ /**
+ * Solves directly at the coarsest level
+ */
+ private void directSolve() {
+ int k = m - 1;
+ u[k].set(f[k]);
+ DenseMatrix U = new DenseMatrix(u[k], false);
+
+ if (transpose)
+ lu.transSolve(U);
+ else
+ lu.solve(U);
+ }
+
+ /**
+ * Applies the relaxation scheme at the given level
+ *
+ * @param k
+ * Multigrid level
+ */
+ private void preRelax(int k) {
+ for (int i = 0; i < nu1; ++i)
+ if (transpose)
+ preM[k].transApply(f[k], u[k]);
+ else
+ preM[k].apply(f[k], u[k]);
+ }
+
+ /**
+ * Applies the relaxation scheme at the given level
+ *
+ * @param k
+ * Multigrid level
+ */
+ private void postRelax(int k) {
+ for (int i = 0; i < nu2; ++i)
+ if (transpose)
+ postM[k].transApply(f[k], u[k]);
+ else
+ postM[k].apply(f[k], u[k]);
+ }
+
+ /**
+ * Creates aggregates. These are disjoint sets, each of which represents one
+ * node at a coarser mesh by aggregating together a set of fine nodes
+ */
+ private static class Aggregator {
+
+ /**
+ * The aggregates
+ */
+ private List<Set<Integer>> C;
+
+ /**
+ * Diagonal indices into the sparse matrix
+ */
+ private int[] diagind;
+
+ /**
+ * The strongly coupled node neighborhood of a given node
+ */
+ private List<Set<Integer>> N;
+
+ /**
+ * Creates the aggregates
+ *
+ * @param A
+ * Sparse matrix
+ * @param eps
+ * Tolerance for selecting the strongly coupled node
+ * neighborhoods. Between zero and one.
+ */
+ public Aggregator(CompRowMatrix A, double eps) {
+
+ diagind = findDiagonalIndices(A);
+ N = findNodeNeighborhood(A, diagind, eps);
+
+ /*
+ * Initialization. Remove isolated nodes from the aggregates
+ */
+
+ boolean[] R = createInitialR(A);
+
+ /*
+ * Startup aggregation. Use disjoint strongly coupled neighborhoods
+ * as the initial aggregate approximation
+ */
+
+ C = createInitialAggregates(N, R);
+
+ /*
+ * Enlargment of the aggregates. Add nodes to each aggregate based
+ * on how strongly connected the nodes are to a given aggregate
+ */
+
+ C = enlargeAggregates(C, N, R);
+
+ /*
+ * Handling of the remenants. Put all remaining unallocated nodes
+ * into new aggregates defined by the intersection of N and R
+ */
+
+ C = createFinalAggregates(C, N, R);
+ }
+
+ /**
+ * Gets the aggregates
+ */
+ public List<Set<Integer>> getAggregates() {
+ return C;
+ }
+
+ /**
+ * Returns the matrix diagonal indices. This is a by-product of the
+ * aggregation
+ */
+ public int[] getDiagonalIndices() {
+ return diagind;
+ }
+
+ /**
+ * Returns the strongly coupled node neighborhoods of a given node. This
+ * is a by-product of the aggregation
+ */
+ public List<Set<Integer>> getNodeNeighborhoods() {
+ return N;
+ }
+
+ /**
+ * Finds the diagonal indices of the matrix
+ */
+ private int[] findDiagonalIndices(CompRowMatrix A) {
+ int[] rowptr = A.getRowPointers();
+ int[] colind = A.getColumnIndices();
+
+ int[] diagind = new int[A.numRows()];
+
+ for (int i = 0; i < A.numRows(); ++i) {
+ diagind[i] = no.uib.cipr.matrix.sparse.Arrays.binarySearch(
+ colind, i, rowptr[i], rowptr[i + 1]);
+ if (diagind[i] < 0)
+ throw new RuntimeException(
+ "Matrix is missing a diagonal entry on row "
+ + (i + 1));
+ }
+
+ return diagind;
+ }
+
+ /**
+ * Finds the strongly coupled node neighborhoods
+ */
+ private List<Set<Integer>> findNodeNeighborhood(CompRowMatrix A,
+ int[] diagind, double eps) {
+
+ N = new ArrayList<Set<Integer>>(A.numRows());
+
+ int[] rowptr = A.getRowPointers();
+ int[] colind = A.getColumnIndices();
+ double[] data = A.getData();
+
+ for (int i = 0; i < A.numRows(); ++i) {
+ Set<Integer> Ni = new HashSet<Integer>();
+
+ double aii = data[diagind[i]];
+ for (int j = rowptr[i]; j < rowptr[i + 1]; ++j) {
+ double aij = data[j];
+ double ajj = data[diagind[colind[j]]];
+
+ if (Math.abs(aij) >= eps * Math.sqrt(aii * ajj))
+ Ni.add(colind[j]);
+ }
+
+ N.add(Ni);
+ }
+
+ return N;
+ }
+
+ /**
+ * Creates the initial R-set by including only the connected nodes
+ */
+ private boolean[] createInitialR(CompRowMatrix A) {
+ boolean[] R = new boolean[A.numRows()];
+
+ int[] rowptr = A.getRowPointers();
+ int[] colind = A.getColumnIndices();
+ double[] data = A.getData();
+
+ for (int i = 0; i < A.numRows(); ++i) {
+ boolean hasOffDiagonal = false;
+
+ for (int j = rowptr[i]; j < rowptr[i + 1]; ++j)
+ if (colind[j] != i && data[j] != 0) {
+ hasOffDiagonal = true;
+ break;
+ }
+
+ R[i] = hasOffDiagonal;
+ }
+
+ return R;
+ }
+
+ /**
+ * Creates the initial aggregates
+ */
+ private List<Set<Integer>> createInitialAggregates(
+ List<Set<Integer>> N, boolean[] R) {
+ C = new ArrayList<Set<Integer>>();
+
+ for (int i = 0; i < R.length; ++i) {
+
+ // Skip non-free nodes
+ if (!R[i])
+ continue;
+
+ // See if all nodes in the current N-set are free
+ boolean free = true;
+ for (int j : N.get(i))
+ free &= R[j];
+
+ // Create an aggregate out of N[i]
+ if (free) {
+ C.add(new HashSet<Integer>(N.get(i)));
+ for (int j : N.get(i))
+ R[j] = false;
+ }
+
+ }
+
+ return C;
+ }
+
+ /**
+ * Enlarges the aggregates
+ */
+ private List<Set<Integer>> enlargeAggregates(List<Set<Integer>> C,
+ List<Set<Integer>> N, boolean[] R) {
+
+ // Contains the aggregates each node is coupled to
+ List<List<Integer>> belong = new ArrayList<List<Integer>>(R.length);
+ for (int i = 0; i < R.length; ++i)
+ belong.add(new ArrayList<Integer>());
+
+ // Find which aggregate each node is coupled to. This is used for
+ // the intersection between Ni and Ck
+ for (int k = 0; k < C.size(); ++k)
+ for (int j : C.get(k))
+ belong.get(j).add(k);
+
+ // Number of nodes in the intersection between each C and Ni
+ int[] intersect = new int[C.size()];
+
+ for (int i = 0; i < R.length; ++i) {
+
+ // Skip non-free nodes
+ if (!R[i])
+ continue;
+
+ // Find the number of nodes intersecting Ni and every C, and
+ // keep a track on the largest overlap
+ Arrays.fill(intersect, 0);
+ int largest = 0, maxValue = 0;
+ for (int j : N.get(i))
+
+ // The k-index is to an aggregate coupled to node j
+ for (int k : belong.get(j)) {
+ intersect[k]++;
+ if (intersect[k] > maxValue) {
+ largest = k;
+ maxValue = intersect[largest];
+ }
+ }
+
+ // Add the node to the proper C-set, and mark it as used
+ // Also, check if the node actually does couple to a set
+ if (maxValue > 0) {
+ R[i] = false;
+ C.get(largest).add(i);
+ }
+ }
+
+ return C;
+ }
+
+ /**
+ * Creates final aggregates from the remaining unallocated nodes
+ */
+ private List<Set<Integer>> createFinalAggregates(List<Set<Integer>> C,
+ List<Set<Integer>> N, boolean[] R) {
+
+ for (int i = 0; i < R.length; ++i) {
+
+ // Skip non-free nodes
+ if (!R[i])
+ continue;
+
+ // Create new aggregate from the nodes in N[i] which are free
+ Set<Integer> Cn = new HashSet<Integer>();
+ for (int j : N.get(i))
+ if (R[j]) {
+ R[j] = false;
+ Cn.add(j);
+ }
+
+ if (!Cn.isEmpty())
+ C.add(Cn);
+ }
+
+ return C;
+ }
+ }
+
+ /**
+ * Creates interpolation (prolongation) operators using based on the
+ * aggregates. Can optionally smooth the aggregates
+ */
+ private static class Interpolator {
+
+ /**
+ * The Galerkin coarse-space operator
+ */
+ private CompRowMatrix Ac;
+
+ /**
+ * The interpolation (prolongation) matrix
+ */
+ private CompColMatrix I;
+
+ /**
+ * Creates the interpolation (prolongation) and Galerkin operators
+ *
+ * @param aggregator
+ * Aggregates
+ * @param A
+ * Matrix
+ * @param omega
+ * Jacobi damping parameter between zero and one. If zero, no
+ * smoothing is performed, and a faster algorithm for forming
+ * the Galerkin operator will be used.
+ */
+ public Interpolator(Aggregator aggregator, CompRowMatrix A, double omega) {
+ List<Set<Integer>> C = aggregator.getAggregates();
+ List<Set<Integer>> N = aggregator.getNodeNeighborhoods();
+ int[] diagind = aggregator.getDiagonalIndices();
+
+ // Create the tentative prolongation, in compressed form
+ int[] pt = createTentativeProlongation(C, A.numRows());
+
+ /*
+ * Apply Jacobi smoothing to the prolongator
+ */
+
+ if (omega != 0) {
+
+ // Smooth the operator by a damped Jacobi method
+ List<Map<Integer, Double>> P = createSmoothedProlongation(C, N,
+ A, diagind, omega, pt);
+
+ // Form a compressed column storage for the operator
+ I = createInterpolationMatrix(P, A.numRows());
+
+ // Create the Galerkin operator using a slow method
+ Ac = createGalerkinSlow(I, A);
+ }
+
+ /*
+ * Use the aggregates as-is
+ */
+
+ else {
+
+ // Create the Galerkin operator using a fast method
+ Ac = createGalerkinFast(A, pt, C.size());
+
+ // Form an explicit interpolation operator
+ I = createInterpolationMatrix(pt, C.size());
+ }
+ }
+
+ /**
+ * Creates the tentative prolongation operator. Since the columns are
+ * all disjoint, and its entries are binary, it is possible to store it
+ * in a single array. Its length equals the number of fine nodes, and
+ * the entries are the indices to the corresponding aggregate (C-set).
+ */
+ private int[] createTentativeProlongation(List<Set<Integer>> C, int n) {
+ int[] pt = new int[n];
+ Arrays.fill(pt, -1);
+
+ for (int i = 0; i < C.size(); ++i)
+ for (int j : C.get(i))
+ pt[j] = i;
+
+ return pt;
+ }
+
+ /**
+ * Creates the Galerkin operator using the assumption of disjoint
+ * (non-smoothed) aggregates
+ */
+ private CompRowMatrix createGalerkinFast(CompRowMatrix A, int[] pt,
+ int c) {
+ int n = pt.length;
+
+ FlexCompRowMatrix Ac = new FlexCompRowMatrix(c, c);
+
+ int[] rowptr = A.getRowPointers();
+ int[] colind = A.getColumnIndices();
+ double[] data = A.getData();
+
+ for (int i = 0; i < n; ++i)
+ if (pt[i] != -1)
+ for (int j = rowptr[i]; j < rowptr[i + 1]; ++j)
+ if (pt[colind[j]] != -1)
+ Ac.add(pt[i], pt[colind[j]], data[j]);
+
+ return new CompRowMatrix(Ac);
+ }
+
+ /**
+ * Creates the interpolation (prolongation) matrix based on the smoothed
+ * aggregates
+ */
+ private CompColMatrix createInterpolationMatrix(
+ List<Map<Integer, Double>> P, int n) {
+
+ // Determine the sparsity pattern of I
+ int c = P.size();
+ int[][] nz = new int[c][];
+ for (int j = 0; j < c; ++j) {
+
+ Map<Integer, Double> Pj = P.get(j);
+ nz[j] = new int[Pj.size()];
+
+ int l = 0;
+ for (int k : Pj.keySet())
+ nz[j][l++] = k;
+ }
+
+ I = new CompColMatrix(n, c, nz);
+
+ // Populate it with numerical entries
+ for (int j = 0; j < c; ++j) {
+
+ Map<Integer, Double> Pj = P.get(j);
+
+ for (Map.Entry<Integer, Double> e : Pj.entrySet())
+ I.set(e.getKey(), j, e.getValue());
+ }
+
+ return I;
+ }
+
+ /**
+ * Creates the interpolation (prolongation) matrix based on the
+ * non-smoothed aggregates
+ */
+ private CompColMatrix createInterpolationMatrix(int[] pt, int c) {
+ FlexCompColMatrix If = new FlexCompColMatrix(pt.length, c);
+
+ for (int i = 0; i < pt.length; ++i)
+ if (pt[i] != -1)
+ If.set(i, pt[i], 1);
+
+ return new CompColMatrix(If);
+ }
+
+ /**
+ * Gets the interpolation (prolongation) operator
+ */
+ public CompColMatrix getInterpolationOperator() {
+ return I;
+ }
+
+ /**
+ * Creates the smoothes interpolation (prolongation) operator by a
+ * single sweep of the damped Jacobi method
+ */
+ private List<Map<Integer, Double>> createSmoothedProlongation(
+ List<Set<Integer>> C, List<Set<Integer>> N, CompRowMatrix A,
+ int[] diagind, double omega, int[] pt) {
+
+ int n = A.numRows(), c = C.size();
+
+ // Allocate the interpolation (prolongation) operator
+ // It is stored by columns, so the maps take row-indices as keys
+ List<Map<Integer, Double>> P = new ArrayList<Map<Integer, Double>>(
+ c);
+ for (int i = 0; i < c; ++i)
+ P.add(new HashMap<Integer, Double>());
+
+ int[] rowptr = A.getRowPointers();
+ int[] colind = A.getColumnIndices();
+ double[] data = A.getData();
+
+ double[] dot = new double[c];
+
+ // Apply the damped Jacobi smoother
+ for (int i = 0; i < n; ++i) {
+
+ if (pt[i] == -1)
+ continue;
+
+ Arrays.fill(dot, 0);
+ Set<Integer> Ni = N.get(i);
+
+ // Calculate A*Pt, except for the diagonal
+ double weakAij = 0;
+ for (int j = rowptr[i]; j < rowptr[i + 1]; ++j) {
+
+ if (pt[colind[j]] == -1)
+ continue;
+
+ double aij = data[j];
+
+ // Off-diagonal, include only strong couplings, and add the
+ // weak couplings to the diagonal
+ if (aij != 0 && !Ni.contains(colind[j])) {
+ weakAij += aij;
+ continue;
+ }
+
+ dot[pt[colind[j]]] += aij;
+ }
+
+ // Subtract the weak couplings from the diagonal part of A*Pt
+ dot[pt[i]] -= weakAij;
+
+ // Scale by omega and the inverse of the diagonal (damping)
+ double scale = -omega / data[diagind[i]];
+ for (int j = 0; j < dot.length; ++j)
+ dot[j] *= scale;
+
+ // Set to (I-omega*D^{-1}*A)*Pt
+ dot[pt[i]]++;
+
+ // This has formed a whole row of P=(I-omega*D^{-1}*A)*Pt
+ // Store the non-zeros into the sparse structure
+ for (int j = 0; j < dot.length; ++j)
+ if (dot[j] != 0)
+ P.get(j).put(i, dot[j]);
+ }
+
+ return P;
+ }
+
+ /**
+ * Creates the entries of the Galerkin operator
+ * <code>Ac = I<sup>T</sup> A I</code>. This is a very
+ * time-consuming operation
+ */
+ private CompRowMatrix createGalerkinSlow(CompColMatrix I,
+ CompRowMatrix A) {
+ int n = I.numRows(), c = I.numColumns();
+ FlexCompRowMatrix Ac = new FlexCompRowMatrix(c, c);
+
+ double[] aiCol = new double[n];
+ double[] iCol = new double[n];
+ DenseVector aiV = new DenseVector(aiCol, false);
+ DenseVector iV = new DenseVector(iCol, false);
+ double[] itaiCol = new double[c];
+ DenseVector itaiV = new DenseVector(itaiCol, false);
+
+ int[] colptr = I.getColumnPointers();
+ int[] rowind = I.getRowIndices();
+ double[] Idata = I.getData();
+
+ for (int k = 0; k < c; ++k) {
+
+ // Expand column 'k' of I to dense storage
+ iV.zero();
+ for (int i = colptr[k]; i < colptr[k + 1]; ++i)
+ iCol[rowind[i]] = Idata[i];
+
+ // Form column 'k' of A*I
+ A.mult(iV, aiV);
+
+ // Form column 'k' of I'*A*I
+ I.transMult(aiV, itaiV);
+
+ // Store non-zeros into Ac
+ for (int i = 0; i < c; ++i)
+ if (itaiCol[i] != 0)
+ Ac.set(i, k, itaiCol[i]);
+ }
+
+ return new CompRowMatrix(Ac);
+ }
+
+ /**
+ * Gets the Galerkin operator
+ */
+ public CompRowMatrix getGalerkinOperator() {
+ return Ac;
+ }
+
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterationMonitor.java b/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterationMonitor.java
new file mode 100644
index 0000000..05683ad
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterationMonitor.java
@@ -0,0 +1,126 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Vector.Norm;
+
+/**
+ * Partial implementation of an iteration reporter
+ */
+public abstract class AbstractIterationMonitor implements IterationMonitor {
+
+ /**
+ * Iteration number
+ */
+ protected int iter;
+
+ /**
+ * Vector-norm
+ */
+ protected Norm normType;
+
+ /**
+ * Iteration reporter
+ */
+ protected IterationReporter reporter;
+
+ /**
+ * Current residual
+ */
+ protected double residual;
+
+ /**
+ * Constructor for AbstractIterationMonitor. Default norm is the 2-norm with
+ * no iteration reporting.
+ */
+ public AbstractIterationMonitor() {
+ normType = Norm.Two;
+ reporter = new NoIterationReporter();
+ }
+
+ public void setFirst() {
+ iter = 0;
+ }
+
+ public boolean isFirst() {
+ return iter == 0;
+ }
+
+ public void next() {
+ iter++;
+ }
+
+ public int iterations() {
+ return iter;
+ }
+
+ public boolean converged(Vector r, Vector x)
+ throws IterativeSolverNotConvergedException {
+ return converged(r.norm(normType), x);
+ }
+
+ public boolean converged(double r, Vector x)
+ throws IterativeSolverNotConvergedException {
+ reporter.monitor(r, x, iter);
+ this.residual = r;
+ return convergedI(r, x);
+ }
+
+ public boolean converged(double r)
+ throws IterativeSolverNotConvergedException {
+ reporter.monitor(r, iter);
+ this.residual = r;
+ return convergedI(r);
+ }
+
+ protected abstract boolean convergedI(double r, Vector x)
+ throws IterativeSolverNotConvergedException;
+
+ protected abstract boolean convergedI(double r)
+ throws IterativeSolverNotConvergedException;
+
+ public boolean converged(Vector r)
+ throws IterativeSolverNotConvergedException {
+ return converged(r.norm(normType));
+ }
+
+ public Norm getNormType() {
+ return normType;
+ }
+
+ public void setNormType(Norm normType) {
+ this.normType = normType;
+ }
+
+ public IterationReporter getIterationReporter() {
+ return reporter;
+ }
+
+ public void setIterationReporter(IterationReporter monitor) {
+ this.reporter = monitor;
+ }
+
+ public double residual() {
+ return residual;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterativeSolver.java b/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterativeSolver.java
new file mode 100644
index 0000000..34db812
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/AbstractIterativeSolver.java
@@ -0,0 +1,98 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Partial implementation of an iterative solver
+ */
+public abstract class AbstractIterativeSolver implements IterativeSolver {
+
+ /**
+ * Preconditioner to use
+ */
+ protected Preconditioner M;
+
+ /**
+ * Iteration monitor
+ */
+ protected IterationMonitor iter;
+
+ /**
+ * Constructor for AbstractIterativeSolver. Does not use preconditioning,
+ * and uses the default linear iteration object.
+ */
+ public AbstractIterativeSolver() {
+ M = new IdentityPreconditioner();
+ iter = new DefaultIterationMonitor();
+ }
+
+ public void setPreconditioner(Preconditioner M) {
+ this.M = M;
+ }
+
+ public Preconditioner getPreconditioner() {
+ return M;
+ }
+
+ public IterationMonitor getIterationMonitor() {
+ return iter;
+ }
+
+ public void setIterationMonitor(IterationMonitor iter) {
+ this.iter = iter;
+ }
+
+ /**
+ * Checks sizes of input data for {@link #solve(Matrix, Vector, Vector)}.
+ * Throws an exception if the sizes does not match.
+ */
+ protected void checkSizes(Matrix A, Vector b, Vector x) {
+ if (!A.isSquare())
+ throw new IllegalArgumentException("!A.isSquare()");
+ if (b.size() != A.numRows())
+ throw new IllegalArgumentException("b.size() != A.numRows()");
+ if (b.size() != x.size())
+ throw new IllegalArgumentException("b.size() != x.size()");
+ }
+
+ /**
+ * Identity preconditioner which does nothing
+ */
+ private static class IdentityPreconditioner implements Preconditioner {
+
+ public Vector apply(Vector b, Vector x) {
+ return x.set(b);
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ return x.set(b);
+ }
+
+ public void setMatrix(Matrix A) {
+ // nothing to do
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/ArpackSym.java b/src/main/java/no/uib/cipr/matrix/sparse/ArpackSym.java
new file mode 100644
index 0000000..19ff7f4
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/ArpackSym.java
@@ -0,0 +1,156 @@
+package no.uib.cipr.matrix.sparse;
+
+import com.github.fommil.netlib.ARPACK;
+import lombok.extern.java.Log;
+import no.uib.cipr.matrix.*;
+import org.netlib.util.doubleW;
+import org.netlib.util.intW;
+
+import java.util.Comparator;
+import java.util.Map;
+import java.util.TreeMap;
+
+/**
+ * Uses ARPACK to partially solve symmetric eigensystems
+ * (ARPACK is designed to compute a subset of eigenvalues/eigenvectors).
+ *
+ * @author Sam Halliday
+ */
+ at Log
+public class ArpackSym {
+
+ public enum Ritz {
+ /**
+ * compute the NEV largest (algebraic) eigenvalues.
+ */
+ LA,
+ /**
+ * compute the NEV smallest (algebraic) eigenvalues.
+ */
+ SA,
+ /**
+ * compute the NEV largest (in magnitude) eigenvalues.
+ */
+ LM,
+ /**
+ * compute the NEV smallest (in magnitude) eigenvalues.
+ */
+ SM,
+ /**
+ * compute NEV eigenvalues, half from each end of the spectrum
+ */
+ BE
+ }
+
+ private final ARPACK arpack = ARPACK.getInstance();
+
+ private static final double TOL = 0.0001;
+
+ private static final boolean EXPENSIVE_CHECKS = true;
+
+ private final Matrix matrix;
+
+
+ public ArpackSym(Matrix matrix) {
+ if (!matrix.isSquare())
+ throw new IllegalArgumentException("matrix must be square");
+ if (EXPENSIVE_CHECKS)
+ for (MatrixEntry entry : matrix) {
+ if (entry.get() != matrix.get(entry.column(), entry.row()))
+ throw new IllegalArgumentException("matrix must be symmetric");
+ }
+ this.matrix = matrix;
+ }
+
+
+ /**
+ * Solve the eigensystem for the number of eigenvalues requested.
+ * <p>
+ * NOTE: The references to the eigenvectors will keep alive a reference to
+ * a {@code nev * n} double array, so use the {@code copy()} method to free
+ * it up if only a subset is required.
+ *
+ * @param eigenvalues
+ * @param ritz preference for solutions
+ * @return a map from eigenvalues to corresponding eigenvectors.
+ */
+ public Map<Double, DenseVectorSub> solve(int eigenvalues, Ritz ritz) {
+ if (eigenvalues <= 0)
+ throw new IllegalArgumentException(eigenvalues + " <= 0");
+ if (eigenvalues >= matrix.numColumns())
+ throw new IllegalArgumentException(eigenvalues + " >= " + (matrix.numColumns()));
+
+ int n = matrix.numRows();
+ intW nev = new intW(eigenvalues);
+
+ int ncv = Math.min(2 * eigenvalues, n);
+
+ String bmat = "I";
+ String which = ritz.name();
+ doubleW tol = new doubleW(TOL);
+ intW info = new intW(0);
+ int[] iparam = new int[11];
+ iparam[0] = 1;
+ iparam[2] = 300;
+ iparam[6] = 1;
+ intW ido = new intW(0);
+
+ // used for initial residual (if info != 0)
+ // and eventually the output residual
+ double[] resid = new double[n];
+ // Lanczos basis vectors
+ double[] v = new double[n * ncv];
+ // Arnoldi reverse communication
+ double[] workd = new double[3 * n];
+ // private work array
+ double[] workl = new double[ncv * (ncv + 8)];
+ int[] ipntr = new int[11];
+
+ int i = 0;
+ while (true) {
+ i++;
+ arpack.dsaupd(ido, bmat, n, which, nev.val, tol, resid, ncv, v, n, iparam, ipntr, workd, workl, workl.length, info);
+ if (ido.val == 99) break;
+ if (ido.val != -1 && ido.val != 1) throw new IllegalStateException("ido = " + ido.val);
+ // could be refactored to handle the other types of mode
+ av(workd, ipntr[0] - 1, ipntr[1] - 1);
+ }
+
+ ArpackSym.log.fine(i + " iterations for " + n);
+
+ if (info.val != 0) throw new IllegalStateException("info = " + info.val);
+
+ double[] d = new double[nev.val];
+ boolean[] select = new boolean[ncv];
+ double[] z = java.util.Arrays.copyOfRange(v, 0, nev.val * n);
+
+ arpack.dseupd(true, "A", select, d, z, n, 0, bmat, n, which, nev, TOL, resid, ncv, v, n, iparam, ipntr, workd, workl, workl.length, info);
+ if (info.val != 0) throw new IllegalStateException("info = " + info.val);
+
+ int computed = iparam[4];
+ ArpackSym.log.fine("computed " + computed + " eigenvalues");
+
+ Map<Double, DenseVectorSub> solution = new TreeMap<Double, DenseVectorSub>(new Comparator<Double>() {
+ @Override
+ public int compare(Double o1, Double o2) {
+ // highest first
+ return Double.compare(o2, o1);
+ }
+ });
+ DenseVector eigenvectors = new DenseVector(z, false);
+ for (i = 0; i < computed; i++) {
+ double eigenvalue = d[i];
+ DenseVectorSub eigenvector = new DenseVectorSub(eigenvectors, i * n, n);
+ solution.put(eigenvalue, eigenvector);
+ }
+
+ return solution;
+ }
+
+ private void av(double[] work, int input_offset, int output_offset) {
+ DenseVector w = new DenseVector(work, false);
+ Vector x = new DenseVectorSub(w, input_offset, matrix.numColumns());
+ Vector y = new DenseVectorSub(w, output_offset, matrix.numColumns());
+ matrix.mult(x, y);
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/Arrays.java b/src/main/java/no/uib/cipr/matrix/sparse/Arrays.java
new file mode 100644
index 0000000..ee29aa1
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/Arrays.java
@@ -0,0 +1,175 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+/**
+ * Array utilities. Complements <code>java.util.Arrays</code>
+ * @deprecated java.utils.Arrays and Google Guava provide this functionality nowadays.
+ */
+ at Deprecated
+class Arrays {
+
+ private Arrays() {
+ // No need to instantiate
+ }
+
+ /**
+ * Searches for a key in a sorted array, and returns an index to an element
+ * which is greater than or equal key.
+ *
+ * @param index
+ * Sorted array of integers
+ * @param key
+ * Search for something equal or greater
+ * @param begin
+ * Start posisiton in the index
+ * @param end
+ * One past the end position in the index
+ * @return end if nothing greater or equal was found, else an index
+ * satisfying the search criteria
+ */
+ public static int binarySearchGreater(int[] index, int key, int begin,
+ int end) {
+ return binarySearchInterval(index, key, begin, end, true);
+ }
+
+ /**
+ * Searches for a key in a sorted array, and returns an index to an element
+ * which is greater than or equal key.
+ *
+ * @param index
+ * Sorted array of integers
+ * @param key
+ * Search for something equal or greater
+ * @return index.length if nothing greater or equal was found, else an index
+ * satisfying the search criteria
+ */
+ public static int binarySearchGreater(int[] index, int key) {
+ return binarySearchInterval(index, key, 0, index.length, true);
+ }
+
+ /**
+ * Searches for a key in a sorted array, and returns an index to an element
+ * which is smaller than or equal key.
+ *
+ * @param index
+ * Sorted array of integers
+ * @param key
+ * Search for something equal or greater
+ * @param begin
+ * Start posisiton in the index
+ * @param end
+ * One past the end position in the index
+ * @return begin-1 if nothing smaller or equal was found, else an index
+ * satisfying the search criteria
+ */
+ public static int binarySearchSmaller(int[] index, int key, int begin,
+ int end) {
+ return binarySearchInterval(index, key, begin, end, false);
+ }
+
+ /**
+ * Searches for a key in a sorted array, and returns an index to an element
+ * which is smaller than or equal key.
+ *
+ * @param index
+ * Sorted array of integers
+ * @param key
+ * Search for something equal or greater
+ * @return -1 if nothing smaller or equal was found, else an index
+ * satisfying the search criteria
+ */
+ public static int binarySearchSmaller(int[] index, int key) {
+ return binarySearchInterval(index, key, 0, index.length, false);
+ }
+
+ /**
+ * Searches for a key in a subset of a sorted array.
+ *
+ * @param index
+ * Sorted array of integers
+ * @param key
+ * Key to search for
+ * @param begin
+ * Start posisiton in the index
+ * @param end
+ * One past the end position in the index
+ * @return Integer index to key. -1 if not found
+ */
+ public static int binarySearch(int[] index, int key, int begin, int end) {
+ return java.util.Arrays.binarySearch(index, begin, end, key);
+ }
+
+ private static int binarySearchInterval(int[] index, int key, int begin,
+ int end, boolean greater) {
+
+ // Zero length array?
+ if (begin == end)
+ if (greater)
+ return end;
+ else
+ return begin - 1;
+
+ end--; // Last index
+ int mid = (end + begin) >> 1;
+
+ // The usual binary search
+ while (begin <= end) {
+ mid = (end + begin) >> 1;
+
+ if (index[mid] < key)
+ begin = mid + 1;
+ else if (index[mid] > key)
+ end = mid - 1;
+ else
+ return mid;
+ }
+
+ // No direct match, but an inf/sup was found
+ if ((greater && index[mid] >= key) || (!greater && index[mid] <= key))
+ return mid;
+ // No inf/sup, return at the end of the array
+ else if (greater)
+ return mid + 1; // One past end
+ else
+ return mid - 1; // One before start
+ }
+
+ /**
+ * Finds the number of repeated entries
+ *
+ * @param num
+ * Maximum index value
+ * @param ind
+ * Indices to check for repetitions
+ * @return Array of length <code>num</code> with the number of repeated
+ * indices of <code>ind</code>
+ */
+ public static int[] bandwidth(int num, int[] ind) {
+ int[] nz = new int[num];
+
+ for (int i = 0; i < ind.length; ++i)
+ nz[ind[i]]++;
+
+ return nz;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/BiCG.java b/src/main/java/no/uib/cipr/matrix/sparse/BiCG.java
new file mode 100644
index 0000000..e307e21
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/BiCG.java
@@ -0,0 +1,104 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * BiCG solver. BiCG solves the unsymmetric linear system <code>Ax = b</code>
+ * using the Preconditioned BiConjugate Gradient method.
+ *
+ * @author Templates
+ */
+public class BiCG extends AbstractIterativeSolver {
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector z, p, q, r, ztilde, ptilde, qtilde, rtilde;
+
+ /**
+ * Constructor for BiCG. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public BiCG(Vector template) {
+ z = template.copy();
+ p = template.copy();
+ q = template.copy();
+ r = template.copy();
+ ztilde = template.copy();
+ ptilde = template.copy();
+ qtilde = template.copy();
+ rtilde = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double rho_1 = 1, rho_2 = 1, alpha = 1, beta = 1;
+
+ A.multAdd(-1, x, r.set(b));
+ rtilde.set(r);
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ M.apply(r, z);
+ M.transApply(rtilde, ztilde);
+ rho_1 = z.dot(rtilde);
+
+ if (rho_1 == 0.)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "rho", iter);
+
+ if (iter.isFirst()) {
+ p.set(z);
+ ptilde.set(ztilde);
+ } else {
+ beta = rho_1 / rho_2;
+ p.scale(beta).add(z);
+ ptilde.scale(beta).add(ztilde);
+ }
+
+ A.mult(p, q);
+ A.transMult(ptilde, qtilde);
+
+ alpha = rho_1 / ptilde.dot(q);
+ x.add(alpha, p);
+ r.add(-alpha, q);
+ rtilde.add(-alpha, qtilde);
+
+ rho_2 = rho_1;
+ }
+
+ return x;
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/BiCGstab.java b/src/main/java/no/uib/cipr/matrix/sparse/BiCGstab.java
new file mode 100644
index 0000000..9ef857f
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/BiCGstab.java
@@ -0,0 +1,118 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * BiCG stablized solver. BiCGstab solves the unsymmetric linear system
+ * <code>Ax = b</code> using the Preconditioned BiConjugate Gradient
+ * Stabilized method
+ *
+ * @author Templates
+ */
+public class BiCGstab extends AbstractIterativeSolver {
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector p, s, phat, shat, t, v, temp, r, rtilde;
+
+ /**
+ * Constructor for BiCGstab. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public BiCGstab(Vector template) {
+ p = template.copy();
+ s = template.copy();
+ phat = template.copy();
+ shat = template.copy();
+ t = template.copy();
+ v = template.copy();
+ temp = template.copy();
+ r = template.copy();
+ rtilde = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double rho_1 = 1, rho_2 = 1, alpha = 1, beta = 1, omega = 1;
+
+ A.multAdd(-1, x, r.set(b));
+ rtilde.set(r);
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ rho_1 = rtilde.dot(r);
+
+ if (rho_1 == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "rho", iter);
+
+ if (omega == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "omega", iter);
+
+ if (iter.isFirst())
+ p.set(r);
+ else {
+ beta = (rho_1 / rho_2) * (alpha / omega);
+
+ // temp = p - omega * v
+ temp.set(-omega, v).add(p);
+
+ // p = r + beta * temp = r + beta * (p - omega * v)
+ p.set(r).add(beta, temp);
+ }
+
+ M.apply(p, phat);
+ A.mult(phat, v);
+ alpha = rho_1 / rtilde.dot(v);
+ s.set(r).add(-alpha, v);
+
+ x.add(alpha, phat);
+ if (iter.converged(s, x))
+ return x;
+
+ M.apply(s, shat);
+ A.mult(shat, t);
+ omega = t.dot(s) / t.dot(t);
+ x.add(omega, shat);
+ r.set(s).add(-omega, t);
+
+ rho_2 = rho_1;
+ }
+
+ return x;
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/CG.java b/src/main/java/no/uib/cipr/matrix/sparse/CG.java
new file mode 100644
index 0000000..7e22720
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/CG.java
@@ -0,0 +1,89 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Conjugate Gradients solver. CG solves the symmetric positive definite linear
+ * system <code>Ax=b</code> using the Conjugate Gradient method.
+ *
+ * @author Templates
+ */
+public class CG extends AbstractIterativeSolver {
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector p, z, q, r;
+
+ /**
+ * Constructor for CG. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public CG(Vector template) {
+ p = template.copy();
+ z = template.copy();
+ q = template.copy();
+ r = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double alpha = 0, beta = 0, rho = 0, rho_1 = 0;
+
+ A.multAdd(-1, x, r.set(b));
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ M.apply(r, z);
+ rho = r.dot(z);
+
+ if (iter.isFirst())
+ p.set(z);
+ else {
+ beta = rho / rho_1;
+ p.scale(beta).add(z);
+ }
+
+ A.mult(p, q);
+ alpha = rho / p.dot(q);
+
+ x.add(alpha, p);
+ r.add(-alpha, q);
+
+ rho_1 = rho;
+ }
+
+ return x;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/CGS.java b/src/main/java/no/uib/cipr/matrix/sparse/CGS.java
new file mode 100644
index 0000000..b49cc61
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/CGS.java
@@ -0,0 +1,107 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Conjugate Gradients squared solver. CGS solves the unsymmetric linear system
+ * <code>Ax = b</code> using the Conjugate Gradient Squared method
+ *
+ * @author Templates
+ */
+public class CGS extends AbstractIterativeSolver {
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector p, q, u, phat, qhat, vhat, uhat, sum, r, rtilde;
+
+ /**
+ * Constructor for CGS. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public CGS(Vector template) {
+ p = template.copy();
+ q = template.copy();
+ u = template.copy();
+ phat = template.copy();
+ qhat = template.copy();
+ vhat = template.copy();
+ uhat = template.copy();
+ sum = template.copy();
+ r = template.copy();
+ rtilde = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double rho_1 = 0, rho_2 = 0, alpha = 0, beta = 0;
+
+ A.multAdd(-1, x, r.set(b));
+ rtilde.set(r);
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ rho_1 = rtilde.dot(r);
+
+ if (rho_1 == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "rho", iter);
+
+ if (iter.isFirst()) {
+ u.set(r);
+ p.set(u);
+ } else {
+ beta = rho_1 / rho_2;
+ u.set(r).add(beta, q);
+ sum.set(q).add(beta, p);
+ p.set(u).add(beta, sum);
+ }
+
+ M.apply(p, phat);
+ A.mult(phat, vhat);
+ alpha = rho_1 / rtilde.dot(vhat);
+ q.set(-alpha, vhat).add(u);
+
+ M.apply(sum.set(u).add(q), uhat);
+ x.add(alpha, uhat);
+ A.mult(uhat, qhat);
+ r.add(-alpha, qhat);
+
+ rho_2 = rho_1;
+ }
+
+ return x;
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/Chebyshev.java b/src/main/java/no/uib/cipr/matrix/sparse/Chebyshev.java
new file mode 100644
index 0000000..9bdb4bc
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/Chebyshev.java
@@ -0,0 +1,121 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Chebyshev solver. Solves the symmetric positive definite linear system
+ * <code>Ax = b</code> using the Preconditioned Chebyshev Method. Chebyshev
+ * requires an acurate estimate on the bounds of the spectrum of the matrix.
+ *
+ * @author Templates
+ */
+public class Chebyshev extends AbstractIterativeSolver {
+
+ /**
+ * Estimates for the eigenvalue of the matrix
+ */
+ private double eigmin, eigmax;
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector p, z, r, q;
+
+ /**
+ * Constructor for Chebyshev. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified. Eigenvalue estimates
+ * must also be provided
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ * @param eigmin
+ * Smallest eigenvalue. Must be positive
+ * @param eigmax
+ * Largest eigenvalue. Must be positive
+ */
+ public Chebyshev(Vector template, double eigmin, double eigmax) {
+ p = template.copy();
+ z = template.copy();
+ r = template.copy();
+ q = template.copy();
+ setEigenvalues(eigmin, eigmax);
+ }
+
+ /**
+ * Sets the eigenvalue estimates.
+ *
+ * @param eigmin
+ * Smallest eigenvalue. Must be positive
+ * @param eigmax
+ * Largest eigenvalue. Must be positive
+ */
+ public void setEigenvalues(double eigmin, double eigmax) {
+ this.eigmin = eigmin;
+ this.eigmax = eigmax;
+
+ if (eigmin <= 0)
+ throw new IllegalArgumentException("eigmin <= 0");
+ if (eigmax <= 0)
+ throw new IllegalArgumentException("eigmax <= 0");
+ if (eigmin > eigmax)
+ throw new IllegalArgumentException("eigmin > eigmax");
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x) throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double alpha = 0, beta = 0, c = 0, d = 0;
+
+ A.multAdd(-1, x, r.set(b));
+
+ c = (eigmax - eigmin) / 2.0;
+ d = (eigmax + eigmin) / 2.0;
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ M.apply(r, z);
+
+ if (iter.isFirst()) {
+ p.set(z);
+ alpha = 2.0 / d;
+ } else {
+ beta = (alpha * c) / 2.0;
+ beta *= beta;
+ alpha = 1.0 / (d - beta);
+ p.scale(beta).add(z);
+ }
+
+ A.mult(p, q);
+ x.add(alpha, p);
+ r.add(-alpha, q);
+ }
+
+ return x;
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/CompColMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/CompColMatrix.java
new file mode 100644
index 0000000..e577fcf
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/CompColMatrix.java
@@ -0,0 +1,479 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.io.IOException;
+import java.util.ArrayList;
+import java.util.Arrays;
+import java.util.HashSet;
+import java.util.Iterator;
+import java.util.List;
+import java.util.Set;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.io.MatrixInfo;
+import no.uib.cipr.matrix.io.MatrixSize;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+/**
+ * Compressed column storage (CCS) matrix
+ */
+public class CompColMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix data
+ */
+ double[] data;
+
+ /**
+ * Column indices. These are kept sorted within each row.
+ */
+ int[] columnPointer;
+
+ /**
+ * Indices to the start of each row
+ */
+ int[] rowIndex;
+
+ /**
+ * Constructor for CompColMatrix
+ *
+ * @param r
+ * Reader to get sparse matrix from
+ */
+ public CompColMatrix(MatrixVectorReader r) throws IOException {
+ // Start with a zero-sized matrix
+ super(0, 0);
+
+ // Get matrix information. Use the header if present, else just assume
+ // that the matrix stores real numbers without any symmetry
+ MatrixInfo info = null;
+ if (r.hasInfo())
+ info = r.readMatrixInfo();
+ else
+ info = new MatrixInfo(true, MatrixInfo.MatrixField.Real,
+ MatrixInfo.MatrixSymmetry.General);
+
+ // Check that the matrix is in an acceptable format
+ if (info.isPattern())
+ throw new UnsupportedOperationException(
+ "Pattern matrices are not supported");
+ if (info.isDense())
+ throw new UnsupportedOperationException(
+ "Dense matrices are not supported");
+ if (info.isComplex())
+ throw new UnsupportedOperationException(
+ "Complex matrices are not supported");
+
+ // Resize the matrix to correct size
+ MatrixSize size = r.readMatrixSize(info);
+ numRows = size.numRows();
+ numColumns = size.numColumns();
+
+ // Start reading entries
+ int numEntries = size.numEntries();
+ int[] row = new int[numEntries];
+ int[] column = new int[numEntries];
+ double[] entry = new double[numEntries];
+ r.readCoordinate(row, column, entry);
+
+ // Shift the indices from 1 based to 0 based
+ r.add(-1, row);
+ r.add(-1, column);
+
+ // Find the number of entries on each column
+ List<Set<Integer>> cnz = new ArrayList<Set<Integer>>(numColumns);
+ for (int i = 0; i < numColumns; ++i)
+ cnz.add(new HashSet<Integer>());
+
+ for (int i = 0; i < numEntries; ++i)
+ cnz.get(column[i]).add(row[i]);
+
+ // Allocate some more in case of symmetry
+ if (info.isSymmetric() || info.isSkewSymmetric())
+ for (int i = 0; i < numEntries; ++i)
+ if (row[i] != column[i])
+ cnz.get(row[i]).add(column[i]);
+
+ int[][] nz = new int[numColumns][];
+ for (int i = 0; i < numColumns; ++i) {
+ nz[i] = new int[cnz.get(i).size()];
+ int j = 0;
+ for (Integer rowind : cnz.get(i))
+ nz[i][j++] = rowind;
+ }
+
+ // Create the sparse matrix structure
+ construct(nz);
+
+ // Insert the entries
+ for (int i = 0; i < size.numEntries(); ++i)
+ set(row[i], column[i], entry[i]);
+
+ // Put in extra entries from symmetry or skew symmetry
+ if (info.isSymmetric())
+ for (int i = 0; i < numEntries; ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], entry[i]);
+ }
+ else if (info.isSkewSymmetric())
+ for (int i = 0; i < numEntries; ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], -entry[i]);
+ }
+ }
+
+ /**
+ * Constructor for CompColMatrix
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of columns
+ * @param nz
+ * The nonzero column indices on each column
+ */
+ public CompColMatrix(int numRows, int numColumns, int[][] nz) {
+ super(numRows, numColumns);
+ construct(nz);
+ }
+
+ private void construct(int[][] nz) {
+ int nnz = 0;
+ for (int i = 0; i < nz.length; ++i)
+ nnz += nz[i].length;
+
+ columnPointer = new int[numColumns + 1];
+ rowIndex = new int[nnz];
+ data = new double[nnz];
+
+ if (nz.length != numColumns)
+ throw new IllegalArgumentException("nz.length != numColumns");
+
+ for (int i = 1; i <= numColumns; ++i) {
+ columnPointer[i] = columnPointer[i - 1] + nz[i - 1].length;
+
+ for (int j = columnPointer[i - 1], k = 0; j < columnPointer[i]; ++j, ++k) {
+ rowIndex[j] = nz[i - 1][k];
+ if (nz[i - 1][k] < 0 || nz[i - 1][k] >= numRows)
+ throw new IllegalArgumentException("nz[" + (i - 1) + "]["
+ + k + "]=" + nz[i - 1][k]
+ + ", which is not a valid row index");
+ }
+
+ Arrays.sort(rowIndex, columnPointer[i - 1], columnPointer[i]);
+ }
+ }
+
+ private void construct(Matrix A, boolean deep) {
+ if (deep) {
+ if (A instanceof CompColMatrix) {
+ CompColMatrix Ac = (CompColMatrix) A;
+ data = new double[Ac.data.length];
+ columnPointer = new int[Ac.columnPointer.length];
+ rowIndex = new int[Ac.rowIndex.length];
+
+ System.arraycopy(Ac.data, 0, data, 0, data.length);
+ System.arraycopy(Ac.columnPointer, 0, columnPointer, 0,
+ columnPointer.length);
+ System.arraycopy(Ac.rowIndex, 0, rowIndex, 0, rowIndex.length);
+ } else {
+
+ List<Set<Integer>> cnz = new ArrayList<Set<Integer>>(numColumns);
+ for (int i = 0; i < numColumns; ++i)
+ cnz.add(new HashSet<Integer>());
+
+ for (MatrixEntry e : A)
+ cnz.get(e.column()).add(e.row());
+
+ int[][] nz = new int[numColumns][];
+ for (int i = 0; i < numColumns; ++i) {
+ nz[i] = new int[cnz.get(i).size()];
+ int j = 0;
+ for (Integer rowind : cnz.get(i))
+ nz[i][j++] = rowind;
+ }
+
+ construct(nz);
+ set(A);
+
+ }
+ } else {
+ CompColMatrix Ac = (CompColMatrix) A;
+ columnPointer = Ac.getColumnPointers();
+ rowIndex = Ac.getRowIndices();
+ data = Ac.getData();
+ }
+ }
+
+ /**
+ * Constructor for CompColMatrix
+ *
+ * @param A
+ * Copies from this matrix
+ * @param deep
+ * True if the copy is to be deep. If it is a shallow copy,
+ * <code>A</code> must be a <code>CompColMatrix</code>
+ */
+ public CompColMatrix(Matrix A, boolean deep) {
+ super(A);
+ construct(A, deep);
+ }
+
+ /**
+ * Constructor for CompColMatrix
+ *
+ * @param A
+ * Copies from this matrix. The copy will be deep
+ */
+ public CompColMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Returns the column pointers
+ */
+ public int[] getColumnPointers() {
+ return columnPointer;
+ }
+
+ /**
+ * Returns the row indices
+ */
+ public int[] getRowIndices() {
+ return rowIndex;
+ }
+
+ /**
+ * Returns the internal data storage
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ // y = 1/alpha * y
+ y.scale(1 / alpha);
+
+ // y = A*x + y
+ for (int i = 0; i < numColumns; ++i)
+ for (int j = columnPointer[i]; j < columnPointer[i + 1]; ++j)
+ yd[rowIndex[j]] += data[j] * xd[i];
+
+ // y = alpha*y = alpha*A*x + y
+ return y.scale(alpha);
+ }
+
+ @Override
+ public Vector transMult(Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMult(x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < numColumns; ++i) {
+ double dot = 0;
+ for (int j = columnPointer[i]; j < columnPointer[i + 1]; ++j)
+ dot += data[j] * xd[rowIndex[j]];
+ yd[i] = dot;
+ }
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < numColumns; ++i) {
+ double dot = 0;
+ for (int j = columnPointer[i]; j < columnPointer[i + 1]; ++j)
+ dot += data[j] * xd[rowIndex[j]];
+ yd[i] += alpha * dot;
+ }
+
+ return y;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+
+ int index = getIndex(row, column);
+ data[index] = value;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ check(row, column);
+
+ int index = getIndex(row, column);
+ data[index] += value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ check(row, column);
+
+ int index = no.uib.cipr.matrix.sparse.Arrays.binarySearch(rowIndex,
+ row, columnPointer[column], columnPointer[column + 1]);
+
+ if (index >= 0)
+ return data[index];
+ else
+ return 0;
+ }
+
+ /**
+ * Finds the insertion index
+ */
+ private int getIndex(int row, int column) {
+ int i = no.uib.cipr.matrix.sparse.Arrays.binarySearch(rowIndex, row,
+ columnPointer[column], columnPointer[column + 1]);
+
+ if (i != -1 && rowIndex[i] == row)
+ return i;
+ else
+ throw new IndexOutOfBoundsException("Entry (" + (row + 1) + ", "
+ + (column + 1) + ") is not in the matrix structure");
+ }
+
+ @Override
+ public CompColMatrix copy() {
+ return new CompColMatrix(this);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new CompColMatrixIterator();
+ }
+
+ @Override
+ public CompColMatrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ /**
+ * Iterator over a compressed column matrix
+ */
+ private class CompColMatrixIterator implements Iterator<MatrixEntry> {
+
+ private int column, cursor;
+
+ private CompColMatrixEntry entry = new CompColMatrixEntry();
+
+ public CompColMatrixIterator() {
+ // Find first non-empty column
+ nextNonEmptyColumn();
+ }
+
+ /**
+ * Locates the first non-empty column, starting at the current. After
+ * the new column has been found, the cursor is also updated
+ */
+ private void nextNonEmptyColumn() {
+ while (column < numColumns()
+ && columnPointer[column] == columnPointer[column + 1])
+ column++;
+ cursor = columnPointer[column];
+ }
+
+ public boolean hasNext() {
+ return cursor < data.length;
+ }
+
+ public MatrixEntry next() {
+ entry.update(column, cursor);
+
+ // Next position is in the same column
+ if (cursor < columnPointer[column + 1] - 1)
+ cursor++;
+
+ // Next position is at the following (non-empty) column
+ else {
+ column++;
+ nextNonEmptyColumn();
+ }
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Entry of a compressed column matrix
+ */
+ private class CompColMatrixEntry implements MatrixEntry {
+
+ private int column, cursor;
+
+ /**
+ * Updates the entry
+ */
+ public void update(int column, int cursor) {
+ this.column = column;
+ this.cursor = cursor;
+ }
+
+ public int row() {
+ return rowIndex[cursor];
+ }
+
+ public int column() {
+ return column;
+ }
+
+ public double get() {
+ return data[cursor];
+ }
+
+ public void set(double value) {
+ data[cursor] = value;
+ }
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/CompDiagMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/CompDiagMatrix.java
new file mode 100644
index 0000000..d2d6353
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/CompDiagMatrix.java
@@ -0,0 +1,458 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.io.IOException;
+import java.util.Arrays;
+import java.util.Iterator;
+import java.util.Set;
+import java.util.TreeSet;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.io.MatrixInfo;
+import no.uib.cipr.matrix.io.MatrixSize;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+/**
+ * Compressed diagonal storage (CDS) matrix
+ */
+public class CompDiagMatrix extends AbstractMatrix {
+
+ /**
+ * The diagonals
+ */
+ double[][] diag;
+
+ /**
+ * Indices to the start of the diagonal, relative to the main diagonal.
+ * Positive means the number of diagonals shifted up, while negative is the
+ * number of diagonals shifted down
+ */
+ int[] ind;
+
+ /**
+ * Constructor for CompDiagMatrix
+ *
+ * @param r
+ * Reader to get sparse matrix from
+ */
+ public CompDiagMatrix(MatrixVectorReader r) throws IOException {
+ // Start with a zero-sized matrix
+ super(0, 0);
+
+ // Get matrix information. Use the header if present, else use a safe
+ // default
+ MatrixInfo info = null;
+ if (r.hasInfo())
+ info = r.readMatrixInfo();
+ else
+ info = new MatrixInfo(true, MatrixInfo.MatrixField.Real,
+ MatrixInfo.MatrixSymmetry.General);
+ MatrixSize size = r.readMatrixSize(info);
+
+ // Resize the matrix to correct size
+ numRows = size.numRows();
+ numColumns = size.numColumns();
+
+ // Check that the matrix is in an acceptable format
+ if (info.isPattern())
+ throw new UnsupportedOperationException(
+ "Pattern matrices are not supported");
+ if (info.isDense())
+ throw new UnsupportedOperationException(
+ "Dense matrices are not supported");
+ if (info.isComplex())
+ throw new UnsupportedOperationException(
+ "Complex matrices are not supported");
+
+ // Start reading entries
+ int[] row = new int[size.numEntries()], column = new int[size
+ .numEntries()];
+ double[] entry = new double[size.numEntries()];
+ r.readCoordinate(row, column, entry);
+
+ // Shift the indices from 1 based to 0 based
+ r.add(-1, row);
+ r.add(-1, column);
+
+ // Find all the diagonals so that we can preallocate
+ Set<Integer> diags = new TreeSet<Integer>();
+ for (int i = 0; i < size.numEntries(); ++i)
+ diags.add(getDiagonal(row[i], column[i]));
+
+ if (info.isSymmetric() || info.isSkewSymmetric())
+ for (int i = 0; i < size.numEntries(); ++i)
+ if (row[i] != column[i])
+ diags.add(getDiagonal(column[i], row[i]));
+
+ // Convert into an integer array
+ int[] ind = new int[diags.size()];
+ {
+ Integer[] ints = new Integer[diags.size()];
+ diags.toArray(ints);
+ for (int i = 0; i < diags.size(); ++i)
+ ind[i] = ints[i];
+ }
+
+ // Create the structure with preallocation
+ construct(ind);
+
+ // Insert the entries
+ for (int i = 0; i < size.numEntries(); ++i)
+ set(row[i], column[i], entry[i]);
+
+ // Put in missing entries from symmetry or skew symmetry
+ if (info.isSymmetric())
+ for (int i = 0; i < size.numEntries(); ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], entry[i]);
+ }
+ else if (info.isSkewSymmetric())
+ for (int i = 0; i < size.numEntries(); ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], -entry[i]);
+ }
+ }
+
+ /**
+ * Creates a new sparse matrix with the given diagonals preallocated. A
+ * negative index is a subdiagonal, positive is superdiagonal
+ */
+ public CompDiagMatrix(int numRows, int numColumns, int[] diagonal) {
+ super(numRows, numColumns);
+ construct(diagonal);
+ }
+
+ private void construct(int[] diagonal) {
+ diag = new double[diagonal.length][];
+ ind = new int[diagonal.length];
+
+ // Keep the diagonal indices sorted
+ int[] sortedDiagonal = new int[diagonal.length];
+ System.arraycopy(diagonal, 0, sortedDiagonal, 0, diagonal.length);
+ Arrays.sort(sortedDiagonal);
+
+ for (int i = 0; i < diagonal.length; ++i) {
+ ind[i] = sortedDiagonal[i];
+ diag[i] = new double[getDiagSize(sortedDiagonal[i])];
+ }
+ }
+
+ /**
+ * Creates a new sparse matrix without preallocation
+ */
+ public CompDiagMatrix(int numRows, int numColumns) {
+ this(numRows, numColumns, new int[0]);
+ }
+
+ /**
+ * Creates a new sparse matrix copied from the given matrix. Can take a deep
+ * copy or a shallow copy. For the latter, the supplied matrix must be a
+ * CompDiagMatrix. Preallocation is also possible, but is only used for the
+ * deep copy.
+ */
+ public CompDiagMatrix(Matrix A, int[] diagonal, boolean deep) {
+ super(A);
+
+ if (deep) {
+ construct(diagonal);
+ set(A);
+ } else {
+ CompDiagMatrix Ac = (CompDiagMatrix) A;
+ diag = Ac.getDiagonals();
+ ind = Ac.getIndex();
+ }
+ }
+
+ /**
+ * Creates a new sparse matrix copied from the given matrix. Takes a deep
+ * copy, with possibility to specify preallocation
+ */
+ public CompDiagMatrix(Matrix A, int[] diagonal) {
+ this(A, diagonal, true);
+ }
+
+ /**
+ * Creates a new sparse matrix copied from the given matrix. Can take a deep
+ * copy or a shallow copy. For the latter, the supplied matrix must be a
+ * CompDiagMatrix. No preallocation is done
+ */
+ public CompDiagMatrix(Matrix A, boolean deep) {
+ this(A, new int[0], deep);
+ }
+
+ /**
+ * Creates a new sparse matrix copied from the given matrix. Takes a deep
+ * copy without preallocation
+ */
+ public CompDiagMatrix(Matrix A) {
+ this(A, new int[0], true);
+ }
+
+ /**
+ * Returns the internal diagonal storage
+ */
+ public double[][] getDiagonals() {
+ return diag;
+ }
+
+ /**
+ * Returns the diagonal offsets
+ */
+ public int[] getIndex() {
+ return ind;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ check(row, column);
+
+ int diagonal = getCompDiagIndex(row, column);
+
+ diag[diagonal][getOnDiagIndex(row, column)] += value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ check(row, column);
+
+ int diagonal = Arrays.binarySearch(ind, getDiagonal(row, column));
+
+ if (diagonal >= 0)
+ return diag[diagonal][getOnDiagIndex(row, column)];
+ else
+ return 0;
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+
+ int diagonal = getCompDiagIndex(row, column);
+
+ diag[diagonal][getOnDiagIndex(row, column)] = value;
+ }
+
+ private int getDiagonal(int row, int column) {
+ return column - row;
+ }
+
+ private int getOnDiagIndex(int row, int column) {
+ return row > column ? column : row;
+ }
+
+ private int getCompDiagIndex(int row, int column) {
+ int diagonal = getDiagonal(row, column);
+
+ // Check if the diagonal is already present
+ int index = no.uib.cipr.matrix.sparse.Arrays.binarySearchGreater(ind,
+ diagonal);
+ if (index < ind.length && ind[index] == diagonal)
+ return index;
+
+ // Need to allocate new diagonal. Get the diagonal size
+ int size = getDiagSize(diagonal);
+
+ // Allocate new primary structure
+ double[] newDiag = new double[size];
+ double[][] newDiagArray = new double[diag.length + 1][];
+ int[] newInd = new int[ind.length + 1];
+
+ // Move data from the old into the new structure
+ System.arraycopy(ind, 0, newInd, 0, index);
+ System.arraycopy(ind, index, newInd, index + 1, ind.length - index);
+ for (int i = 0; i < index; ++i)
+ newDiagArray[i] = diag[i];
+ for (int i = index; i < diag.length; ++i)
+ newDiagArray[i + 1] = diag[i];
+
+ newInd[index] = diagonal;
+ newDiagArray[index] = newDiag;
+
+ // Update pointers
+ ind = newInd;
+ diag = newDiagArray;
+
+ return index;
+ }
+
+ /**
+ * Finds the size of the requested diagonal to be allocated
+ */
+ private int getDiagSize(int diagonal) {
+ if (diagonal < 0)
+ return Math.min(numRows + diagonal, numColumns);
+ else
+ return Math.min(numRows, numColumns - diagonal);
+ }
+
+ @Override
+ public Matrix copy() {
+ return new CompDiagMatrix(this, ind);
+ }
+
+ @Override
+ public Matrix zero() {
+ for (int i = 0; i < diag.length; ++i)
+ Arrays.fill(diag[i], 0);
+ return this;
+ }
+
+ @Override
+ public Vector mult(Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.mult(x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ y.zero();
+
+ for (int i = 0; i < ind.length; ++i) {
+ int row = ind[i] < 0 ? -ind[i] : 0;
+ int column = ind[i] > 0 ? ind[i] : 0;
+ double[] locDiag = diag[i];
+ for (int j = 0; j < locDiag.length; ++j, ++row, ++column)
+ yd[row] += locDiag[j] * xd[column];
+ }
+
+ return y;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < ind.length; ++i) {
+ int row = ind[i] < 0 ? -ind[i] : 0;
+ int column = ind[i] > 0 ? ind[i] : 0;
+ double[] locDiag = diag[i];
+ for (int j = 0; j < locDiag.length; ++j, ++row, ++column)
+ yd[row] += alpha * locDiag[j] * xd[column];
+ }
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < ind.length; ++i) {
+ int row = ind[i] < 0 ? -ind[i] : 0;
+ int column = ind[i] > 0 ? ind[i] : 0;
+ double[] locDiag = diag[i];
+ for (int j = 0; j < locDiag.length; ++j, ++row, ++column)
+ yd[column] += alpha * locDiag[j] * xd[row];
+ }
+
+ return y;
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new CompDiagMatrixIterator();
+ }
+
+ /**
+ * Iterator over a compressed diagonal matrix
+ */
+ private class CompDiagMatrixIterator implements Iterator<MatrixEntry> {
+
+ private int diagonal, index;
+
+ private CompDiagMatrixEntry entry = new CompDiagMatrixEntry();
+
+ public boolean hasNext() {
+ return diagonal < diag.length;
+ }
+
+ public MatrixEntry next() {
+ entry.update(diagonal, index);
+
+ // Move along current diagonal
+ if (index < diag[diagonal].length - 1)
+ index++;
+
+ // Move to the next diagonal
+ else {
+ diagonal++;
+ index = 0;
+ }
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Entry of a compressed diagonal matrix
+ */
+ private class CompDiagMatrixEntry implements MatrixEntry {
+
+ private int diagonal, index;
+
+ public void update(int diagonal, int index) {
+ this.diagonal = diagonal;
+ this.index = index;
+ }
+
+ public int row() {
+ return index + (ind[diagonal] < 0 ? -ind[diagonal] : 0);
+ }
+
+ public int column() {
+ return index + (ind[diagonal] > 0 ? ind[diagonal] : 0);
+ }
+
+ public double get() {
+ return diag[diagonal][index];
+ }
+
+ public void set(double value) {
+ diag[diagonal][index] = value;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/CompRowMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/CompRowMatrix.java
new file mode 100644
index 0000000..3b2b953
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/CompRowMatrix.java
@@ -0,0 +1,561 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.io.IOException;
+import java.util.ArrayList;
+import java.util.Arrays;
+import java.util.HashSet;
+import java.util.Iterator;
+import java.util.List;
+import java.util.Set;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.io.MatrixInfo;
+import no.uib.cipr.matrix.io.MatrixSize;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+import com.github.fommil.netlib.BLAS;
+
+/**
+ * Compressed row storage (CRS) matrix
+ */
+public class CompRowMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix data
+ */
+ double[] data;
+
+ /**
+ * Column indices. These are kept sorted within each row.
+ */
+ int[] columnIndex;
+
+ /**
+ * Indices to the start of each row
+ */
+ int[] rowPointer;
+
+ /**
+ * Constructor for CompRowMatrix
+ *
+ * @param r
+ * Reader to get sparse matrix from
+ */
+ public CompRowMatrix(MatrixVectorReader r) throws IOException {
+ // Start with a zero-sized matrix
+ super(0, 0);
+
+ // Get matrix information. Use the header if present, else just assume
+ // that the matrix stores real numbers without any symmetry
+ MatrixInfo info = null;
+ if (r.hasInfo())
+ info = r.readMatrixInfo();
+ else
+ info = new MatrixInfo(true, MatrixInfo.MatrixField.Real,
+ MatrixInfo.MatrixSymmetry.General);
+
+ // Check that the matrix is in an acceptable format
+ if (info.isPattern())
+ throw new UnsupportedOperationException(
+ "Pattern matrices are not supported");
+ if (info.isDense())
+ throw new UnsupportedOperationException(
+ "Dense matrices are not supported");
+ if (info.isComplex())
+ throw new UnsupportedOperationException(
+ "Complex matrices are not supported");
+
+ // Resize the matrix to correct size
+ MatrixSize size = r.readMatrixSize(info);
+ numRows = size.numRows();
+ numColumns = size.numColumns();
+
+ // Start reading entries
+ int numEntries = size.numEntries();
+ int[] row = new int[numEntries];
+ int[] column = new int[numEntries];
+ double[] entry = new double[numEntries];
+ r.readCoordinate(row, column, entry);
+
+ // Shift the indices from 1 based to 0 based
+ r.add(-1, row);
+ r.add(-1, column);
+
+ // Find the number of entries on each row
+ List<Set<Integer>> rnz = new ArrayList<Set<Integer>>(numRows);
+ for (int i = 0; i < numRows; ++i)
+ rnz.add(new HashSet<Integer>());
+
+ for (int i = 0; i < numEntries; ++i)
+ rnz.get(row[i]).add(column[i]);
+
+ // Allocate some more in case of symmetry
+ if (info.isSymmetric() || info.isSkewSymmetric())
+ for (int i = 0; i < numEntries; ++i)
+ if (row[i] != column[i])
+ rnz.get(column[i]).add(row[i]);
+
+ int[][] nz = new int[numRows][];
+ for (int i = 0; i < numRows; ++i) {
+ nz[i] = new int[rnz.get(i).size()];
+ int j = 0;
+ for (Integer colind : rnz.get(i))
+ nz[i][j++] = colind;
+ }
+
+ // Create the sparse matrix structure
+ construct(nz);
+
+ // Insert the entries
+ for (int i = 0; i < size.numEntries(); ++i)
+ set(row[i], column[i], entry[i]);
+
+ // Put in extra entries from symmetry or skew symmetry
+ if (info.isSymmetric())
+ for (int i = 0; i < numEntries; ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], entry[i]);
+ }
+ else if (info.isSkewSymmetric())
+ for (int i = 0; i < numEntries; ++i) {
+ if (row[i] != column[i])
+ set(column[i], row[i], -entry[i]);
+ }
+ }
+
+ /**
+ * Constructor for CompRowMatrix
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of columns
+ * @param nz
+ * The nonzero column indices on each row
+ */
+ public CompRowMatrix(int numRows, int numColumns, int[][] nz) {
+ super(numRows, numColumns);
+ construct(nz);
+ }
+
+ private void construct(int[][] nz) {
+ int nnz = 0;
+ for (int i = 0; i < nz.length; ++i)
+ nnz += nz[i].length;
+
+ rowPointer = new int[numRows + 1];
+ columnIndex = new int[nnz];
+ data = new double[nnz];
+
+ if (nz.length != numRows)
+ throw new IllegalArgumentException("nz.length != numRows");
+
+ for (int i = 1; i <= numRows; ++i) {
+ rowPointer[i] = rowPointer[i - 1] + nz[i - 1].length;
+
+ for (int j = rowPointer[i - 1], k = 0; j < rowPointer[i]; ++j, ++k) {
+ columnIndex[j] = nz[i - 1][k];
+ if (nz[i - 1][k] < 0 || nz[i - 1][k] >= numColumns)
+ throw new IllegalArgumentException("nz[" + (i - 1) + "]["
+ + k + "]=" + nz[i - 1][k]
+ + ", which is not a valid column index");
+ }
+
+ Arrays.sort(columnIndex, rowPointer[i - 1], rowPointer[i]);
+ }
+ }
+
+ private void construct(Matrix A, boolean deep) {
+ if (deep) {
+ if (A instanceof CompRowMatrix) {
+ CompRowMatrix Ac = (CompRowMatrix) A;
+ data = new double[Ac.data.length];
+ columnIndex = new int[Ac.columnIndex.length];
+ rowPointer = new int[Ac.rowPointer.length];
+
+ System.arraycopy(Ac.data, 0, data, 0, data.length);
+ System.arraycopy(Ac.columnIndex, 0, columnIndex, 0,
+ columnIndex.length);
+ System.arraycopy(Ac.rowPointer, 0, rowPointer, 0,
+ rowPointer.length);
+ } else {
+
+ List<Set<Integer>> rnz = new ArrayList<Set<Integer>>(numRows);
+ for (int i = 0; i < numRows; ++i)
+ rnz.add(new HashSet<Integer>());
+
+ for (MatrixEntry e : A)
+ rnz.get(e.row()).add(e.column());
+
+ int[][] nz = new int[numRows][];
+ for (int i = 0; i < numRows; ++i) {
+ nz[i] = new int[rnz.get(i).size()];
+ int j = 0;
+ for (Integer colind : rnz.get(i))
+ nz[i][j++] = colind;
+ }
+
+ construct(nz);
+ set(A);
+
+ }
+ } else {
+ CompRowMatrix Ac = (CompRowMatrix) A;
+ columnIndex = Ac.getColumnIndices();
+ rowPointer = Ac.getRowPointers();
+ data = Ac.getData();
+ }
+ }
+
+ /**
+ * Constructor for CompRowMatrix
+ *
+ * @param A
+ * Copies from this matrix
+ * @param deep
+ * True if the copy is to be deep. If it is a shallow copy,
+ * <code>A</code> must be a <code>CompRowMatrix</code>
+ */
+ public CompRowMatrix(Matrix A, boolean deep) {
+ super(A);
+ construct(A, deep);
+ }
+
+ /**
+ * Constructor for CompRowMatrix
+ *
+ * @param A
+ * Copies from this matrix. The copy will be deep
+ */
+ public CompRowMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Returns the column indices
+ */
+ public int[] getColumnIndices() {
+ return columnIndex;
+ }
+
+ /**
+ * Returns the row pointers
+ */
+ public int[] getRowPointers() {
+ return rowPointer;
+ }
+
+ /**
+ * Returns the internal data storage
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ @Override
+ public Matrix mult(Matrix B, Matrix C) {
+ checkMultAdd(B, C);
+ C.zero();
+
+ // optimised a little bit to avoid zeros in rows, but not to
+ // exploit sparsity of matrix B
+ for (int i = 0; i < numRows; ++i) {
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = rowPointer[i]; k < rowPointer[i + 1]; ++k) {
+ dot += data[k] * B.get(columnIndex[k], j);
+ }
+ if (dot != 0) {
+ C.set(i, j, dot);
+ }
+ }
+ }
+ return C;
+ }
+
+ @Override
+ public Vector mult(Vector x, Vector y) {
+ // check dimensions
+ checkMultAdd(x, y);
+ // can't assume this, unfortunately
+ y.zero();
+
+ if (x instanceof DenseVector) {
+ // DenseVector optimisations
+ double[] xd = ((DenseVector) x).getData();
+ for (int i = 0; i < numRows; ++i) {
+ double dot = 0;
+ for (int j = rowPointer[i]; j < rowPointer[i + 1]; j++) {
+ dot += data[j] * xd[columnIndex[j]];
+ }
+ if (dot != 0) {
+ y.set(i, dot);
+ }
+ }
+ return y;
+ }
+ // use sparsity of matrix (not vector), as get(,) is slow
+ // TODO: additional optimisations for mult(ISparseVector, Vector)
+ // note that this would require Sparse BLAS, e.g. BLAS_DUSDOT(,,,,)
+ // @see http://www.netlib.org/blas/blast-forum/chapter3.pdf
+ for (int i = 0; i < numRows; ++i) {
+ double dot = 0;
+ for (int j = rowPointer[i]; j < rowPointer[i + 1]; j++) {
+ dot += data[j] * x.get(columnIndex[j]);
+ }
+ y.set(i, dot);
+ }
+ return y;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ for (int i = 0; i < numRows; ++i) {
+ double dot = 0;
+ for (int j = rowPointer[i]; j < rowPointer[i + 1]; ++j)
+ dot += data[j] * xd[columnIndex[j]];
+ yd[i] += alpha * dot;
+ }
+
+ return y;
+ }
+
+ @Override
+ public Vector transMult(Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMult(x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ y.zero();
+
+ for (int i = 0; i < numRows; ++i)
+ for (int j = rowPointer[i]; j < rowPointer[i + 1]; ++j)
+ yd[columnIndex[j]] += data[j] * xd[i];
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = 1/alpha * y
+ y.scale(1. / alpha);
+
+ // y = A'x + y
+ for (int i = 0; i < numRows; ++i)
+ for (int j = rowPointer[i]; j < rowPointer[i + 1]; ++j)
+ yd[columnIndex[j]] += data[j] * xd[i];
+
+ // y = alpha*y = alpha*A'x + y
+ return y.scale(alpha);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+
+ int index = getIndex(row, column);
+ data[index] = value;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ check(row, column);
+
+ int index = getIndex(row, column);
+ data[index] += value;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ check(row, column);
+
+ int index = no.uib.cipr.matrix.sparse.Arrays.binarySearch(columnIndex,
+ column, rowPointer[row], rowPointer[row + 1]);
+
+ if (index >= 0)
+ return data[index];
+ else
+ return 0;
+ }
+
+ /**
+ * Finds the insertion index
+ */
+ private int getIndex(int row, int column) {
+ int i = no.uib.cipr.matrix.sparse.Arrays.binarySearch(columnIndex,
+ column, rowPointer[row], rowPointer[row + 1]);
+
+ if (i != -1 && columnIndex[i] == column)
+ return i;
+ else
+ throw new IndexOutOfBoundsException("Entry (" + (row + 1) + ", "
+ + (column + 1) + ") is not in the matrix structure");
+ }
+
+ @Override
+ public CompRowMatrix copy() {
+ return new CompRowMatrix(this);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new CompRowMatrixIterator();
+ }
+
+ @Override
+ public CompRowMatrix zero() {
+ Arrays.fill(data, 0);
+ return this;
+ }
+
+ @Override
+ public Matrix set(Matrix B) {
+ if (!(B instanceof CompRowMatrix))
+ return super.set(B);
+
+ checkSize(B);
+
+ CompRowMatrix Bc = (CompRowMatrix) B;
+
+ // Reallocate matrix structure, if necessary
+ if (Bc.columnIndex.length != columnIndex.length
+ || Bc.rowPointer.length != rowPointer.length) {
+ data = new double[Bc.data.length];
+ columnIndex = new int[Bc.columnIndex.length];
+ rowPointer = new int[Bc.rowPointer.length];
+ }
+
+ System.arraycopy(Bc.data, 0, data, 0, data.length);
+ System.arraycopy(Bc.columnIndex, 0, columnIndex, 0, columnIndex.length);
+ System.arraycopy(Bc.rowPointer, 0, rowPointer, 0, rowPointer.length);
+
+ return this;
+ }
+
+ /**
+ * Iterator over a compressed row matrix
+ */
+ private class CompRowMatrixIterator implements Iterator<MatrixEntry> {
+
+ private int row, cursor;
+
+ private CompRowMatrixEntry entry = new CompRowMatrixEntry();
+
+ public CompRowMatrixIterator() {
+ // Find first non-empty row
+ nextNonEmptyRow();
+ }
+
+ /**
+ * Locates the first non-empty row, starting at the current. After the
+ * new row has been found, the cursor is also updated
+ */
+ private void nextNonEmptyRow() {
+ while (row < numRows() && rowPointer[row] == rowPointer[row + 1])
+ row++;
+ cursor = rowPointer[row];
+ }
+
+ public boolean hasNext() {
+ return cursor < data.length;
+ }
+
+ public MatrixEntry next() {
+ entry.update(row, cursor);
+
+ // Next position is in the same row
+ if (cursor < rowPointer[row + 1] - 1)
+ cursor++;
+
+ // Next position is at the following (non-empty) row
+ else {
+ row++;
+ nextNonEmptyRow();
+ }
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Entry of a compressed row matrix
+ */
+ private class CompRowMatrixEntry implements MatrixEntry {
+
+ private int row, cursor;
+
+ /**
+ * Updates the entry
+ */
+ public void update(int row, int cursor) {
+ this.row = row;
+ this.cursor = cursor;
+ }
+
+ public int row() {
+ return row;
+ }
+
+ public int column() {
+ return columnIndex[cursor];
+ }
+
+ public double get() {
+ return data[cursor];
+ }
+
+ public void set(double value) {
+ data[cursor] = value;
+ }
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/DefaultIterationMonitor.java b/src/main/java/no/uib/cipr/matrix/sparse/DefaultIterationMonitor.java
new file mode 100644
index 0000000..988b1c2
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/DefaultIterationMonitor.java
@@ -0,0 +1,164 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Default iteration monitor. This tester checks declares convergence if the
+ * absolute value of the residual norm is sufficiently small, or if the relative
+ * decrease is small. Divergence is decleared if too many iterations are spent,
+ * or the residual has grown too much. NaNs will also cause divergence to be
+ * flagged.
+ */
+public class DefaultIterationMonitor extends AbstractIterationMonitor {
+
+ /**
+ * Initial residual
+ */
+ double initR;
+
+ /**
+ * Relative tolerance
+ */
+ double rtol;
+
+ /**
+ * Absolute tolerance
+ */
+ double atol;
+
+ /**
+ * Divergence tolerance
+ */
+ double dtol;
+
+ /**
+ * Maximum number of iterations
+ */
+ int maxIter;
+
+ /**
+ * Constructor for DefaultIterationMonitor
+ *
+ * @param maxIter
+ * Maximum number of iterations
+ * @param rtol
+ * Relative convergence tolerance (to initial residual)
+ * @param atol
+ * Absolute convergence tolerance
+ * @param dtol
+ * Relative divergence tolerance (to initial residual)
+ */
+ public DefaultIterationMonitor(int maxIter, double rtol, double atol,
+ double dtol) {
+ this.maxIter = maxIter;
+ this.rtol = rtol;
+ this.atol = atol;
+ this.dtol = dtol;
+ }
+
+ /**
+ * Constructor for DefaultIterationMonitor. Default is 100000 iterations at
+ * most, relative tolerance of 1e-5, absolute tolerance of 1e-50 and a
+ * divergence tolerance of 1e+5.
+ */
+ public DefaultIterationMonitor() {
+ this.maxIter = 100000;
+ this.rtol = 1e-5;
+ this.atol = 1e-50;
+ this.dtol = 1e+5;
+ }
+
+ /**
+ * Sets maximum number of iterations to permit
+ *
+ * @param maxIter
+ * Maximum number of iterations
+ */
+ public void setMaxIterations(int maxIter) {
+ this.maxIter = maxIter;
+ }
+
+ /**
+ * Sets the relative tolerance
+ *
+ * @param rtol
+ * Relative convergence tolerance (to initial residual)
+ */
+ public void setRelativeTolerance(double rtol) {
+ this.rtol = rtol;
+ }
+
+ /**
+ * Sets the absolute tolerance
+ *
+ * @param atol
+ * Absolute convergence tolerance
+ */
+ public void setAbsoluteTolerance(double atol) {
+ this.atol = atol;
+ }
+
+ /**
+ * Sets the divergence tolerance
+ *
+ * @param dtol
+ * Relative divergence tolerance (to initial residual)
+ */
+ public void setDivergenceTolerance(double dtol) {
+ this.dtol = dtol;
+ }
+
+ @Override
+ protected boolean convergedI(double r)
+ throws IterativeSolverNotConvergedException {
+ // Store initial residual
+ if (isFirst())
+ initR = r;
+
+ // Check for convergence
+ if (r < Math.max(rtol * initR, atol))
+ return true;
+
+ // Check for divergence
+ if (r > dtol * initR)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Divergence, this);
+ if (iter >= maxIter)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Iterations, this);
+ if (Double.isNaN(r))
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Divergence, this);
+
+ // Neither convergence nor divergence
+ return false;
+ }
+
+ @Override
+ protected boolean convergedI(double r, Vector x)
+ throws IterativeSolverNotConvergedException {
+ return convergedI(r);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/DiagonalPreconditioner.java b/src/main/java/no/uib/cipr/matrix/sparse/DiagonalPreconditioner.java
new file mode 100644
index 0000000..9f88711
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/DiagonalPreconditioner.java
@@ -0,0 +1,78 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Diagonal preconditioner. Uses the inverse of the diagonal as preconditioner
+ */
+public class DiagonalPreconditioner implements Preconditioner {
+
+ /**
+ * This contains the inverse of the diagonal
+ */
+ private double[] invdiag;
+
+ /**
+ * Constructor for DiagonalPreconditioner
+ *
+ * @param n
+ * Problem size (number of rows)
+ */
+ public DiagonalPreconditioner(int n) {
+ invdiag = new double[n];
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ if (!(x instanceof DenseVector) || !(b instanceof DenseVector))
+ throw new IllegalArgumentException("Vector must be DenseVectors");
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] bd = ((DenseVector) b).getData();
+
+ for (int i = 0; i < invdiag.length; ++i)
+ xd[i] = bd[i] * invdiag[i];
+
+ return x;
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ return apply(b, x);
+ }
+
+ public void setMatrix(Matrix A) {
+ if (A.numRows() != invdiag.length)
+ throw new IllegalArgumentException(
+ "Matrix size differs from preconditioner size");
+
+ for (int i = 0; i < invdiag.length; ++i) {
+ invdiag[i] = A.get(i, i);
+ if (invdiag[i] == 0) // Avoid zero-division
+ throw new RuntimeException("Zero diagonal on row " + (i + 1));
+ else
+ invdiag[i] = 1 / invdiag[i];
+ }
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/FlexCompColMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/FlexCompColMatrix.java
new file mode 100644
index 0000000..d6be618
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/FlexCompColMatrix.java
@@ -0,0 +1,253 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.VectorEntry;
+import no.uib.cipr.matrix.sparse.SuperIterator.SuperIteratorEntry;
+
+/**
+ * Matrix stored column-wise into sparse vectors
+ */
+public class FlexCompColMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix data
+ */
+ SparseVector[] colD;
+
+ /**
+ * Constructor for FlexCompColMatrix
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of column
+ */
+ public FlexCompColMatrix(int numRows, int numColumns) {
+ super(numRows, numColumns);
+
+ colD = new SparseVector[numColumns];
+ for (int i = 0; i < numColumns; ++i)
+ colD[i] = new SparseVector(numRows);
+ }
+
+ /**
+ * Constructor for FlexCompColMatrix
+ *
+ * @param A
+ * Matrix to copy contents from
+ * @param deep
+ * True for a deep copy, false for a reference copy. A reference
+ * copy can only be made of an <code>FlexCompColMatrix</code>
+ */
+ public FlexCompColMatrix(Matrix A, boolean deep) {
+ super(A);
+ colD = new SparseVector[numColumns];
+
+ if (deep) {
+ for (int i = 0; i < numColumns; ++i)
+ colD[i] = new SparseVector(numRows);
+ set(A);
+ } else {
+ FlexCompColMatrix Ar = (FlexCompColMatrix) A;
+ for (int i = 0; i < numColumns; ++i)
+ colD[i] = Ar.getColumn(i);
+ }
+ }
+
+ /**
+ * Constructor for FlexCompColMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. The copy will be deep
+ */
+ public FlexCompColMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Returns the given column
+ */
+ public SparseVector getColumn(int i) {
+ return colD[i];
+ }
+
+ /**
+ * Sets the given column equal the passed vector
+ */
+ public void setColumn(int i, SparseVector x) {
+ if (x.size() != numRows)
+ throw new IllegalArgumentException(
+ "New column must be of the same size as existing column");
+ colD[i] = x;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.multAdd(alpha, x, y);
+
+ checkMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData();
+ double[] yd = ((DenseVector) y).getData();
+
+ // y = 1/alpha * y
+ y.scale(1. / alpha);
+
+ // y = A*x + y
+ for (int i = 0; i < numColumns; ++i) {
+ SparseVector v = colD[i];
+ int[] index = v.getIndex();
+ double[] data = v.getData();
+ int length = v.getUsed();
+ for (int j = 0; j < length; ++j)
+ yd[index[j]] += data[j] * xd[i];
+ }
+
+ // y = alpha*y = alpha * A'x + y
+ return y.scale(alpha);
+ }
+
+ @Override
+ public Vector transMultAdd(final double alpha, final Vector x,
+ final Vector y) {
+ checkTransMultAdd(x, y);
+
+ for (int i = 0; i < numColumns; ++i)
+ y.add(i, alpha * colD[i].dot(x));
+
+ return y;
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ colD[column].add(row, value);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ colD[column].set(row, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return colD[column].get(row);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new ColMatrixIterator();
+ }
+
+ @Override
+ public FlexCompColMatrix copy() {
+ return new FlexCompColMatrix(this);
+ }
+
+ @Override
+ public FlexCompColMatrix zero() {
+ for (int i = 0; i < numColumns; ++i)
+ colD[i].zero();
+ return this;
+ }
+
+ /**
+ * Tries to store the matrix as compactly as possible
+ */
+ public void compact() {
+ for (Vector v : colD)
+ ((SparseVector) v).compact();
+ }
+
+ /**
+ * Iterator over a matrix stored vectorwise by columns
+ */
+ private class ColMatrixIterator implements Iterator<MatrixEntry> {
+
+ /**
+ * Iterates over each column vector
+ */
+ private SuperIterator<SparseVector, VectorEntry> iterator = new SuperIterator<SparseVector, VectorEntry>(
+ Arrays.asList(colD));
+
+ /**
+ * Entry returned
+ */
+ private ColMatrixEntry entry = new ColMatrixEntry();
+
+ public boolean hasNext() {
+ return iterator.hasNext();
+ }
+
+ public MatrixEntry next() {
+ SuperIteratorEntry<VectorEntry> se = iterator.next();
+ entry.update(se.index(), se.get());
+ return entry;
+ }
+
+ public void remove() {
+ iterator.remove();
+ }
+
+ }
+
+ /**
+ * Entry of a matrix stored vectorwise by columns
+ */
+ private static class ColMatrixEntry implements MatrixEntry {
+
+ private int column;
+
+ private VectorEntry entry;
+
+ public void update(int column, VectorEntry entry) {
+ this.column = column;
+ this.entry = entry;
+ }
+
+ public int row() {
+ return entry.index();
+ }
+
+ public int column() {
+ return column;
+ }
+
+ public double get() {
+ return entry.get();
+ }
+
+ public void set(double value) {
+ entry.set(value);
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrix.java
new file mode 100644
index 0000000..e4de45c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrix.java
@@ -0,0 +1,267 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.VectorEntry;
+import no.uib.cipr.matrix.sparse.SuperIterator.SuperIteratorEntry;
+
+/**
+ * Matrix stored row-wise into sparse vectors
+ */
+public class FlexCompRowMatrix extends AbstractMatrix {
+
+ /**
+ * Matrix data
+ */
+ SparseVector[] rowD;
+
+ /**
+ * Constructor for FlexCompRowMatrix
+ *
+ * @param numRows
+ * Number of rows
+ * @param numColumns
+ * Number of column
+ */
+ public FlexCompRowMatrix(int numRows, int numColumns) {
+ super(numRows, numColumns);
+
+ rowD = new SparseVector[numRows];
+ for (int i = 0; i < numRows; ++i)
+ rowD[i] = new SparseVector(numColumns);
+ }
+
+ /**
+ * Constructor for FlexCompRowMatrix
+ *
+ * @param A
+ * Matrix to copy contents from
+ * @param deep
+ * True for a deep copy, false for a reference copy. A reference
+ * copy can only be made of an <code>FlexCompRowMatrix</code>
+ */
+ public FlexCompRowMatrix(Matrix A, boolean deep) {
+ super(A);
+ rowD = new SparseVector[numRows];
+
+ if (deep) {
+ for (int i = 0; i < numRows; ++i)
+ rowD[i] = new SparseVector(numColumns);
+ set(A);
+ } else {
+ FlexCompRowMatrix Ar = (FlexCompRowMatrix) A;
+ for (int i = 0; i < numRows; ++i)
+ rowD[i] = Ar.getRow(i);
+ }
+ }
+
+ /**
+ * Constructor for FlexCompRowMatrix
+ *
+ * @param A
+ * Matrix to copy contents from. The copy will be deep
+ */
+ public FlexCompRowMatrix(Matrix A) {
+ this(A, true);
+ }
+
+ /**
+ * Returns the given row
+ */
+ public SparseVector getRow(int i) {
+ return rowD[i];
+ }
+
+ /**
+ * Sets the given row equal the passed vector
+ */
+ public void setRow(int i, SparseVector x) {
+ if (x.size() != numColumns)
+ throw new IllegalArgumentException(
+ "New row must be of the same size as existing row");
+ rowD[i] = x;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ checkMultAdd(x, y);
+
+ for (int i = 0; i < numRows; ++i)
+ y.add(i, alpha * rowD[i].dot(x));
+
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ if (!(x instanceof DenseVector) || !(y instanceof DenseVector))
+ return super.transMultAdd(alpha, x, y);
+
+ checkTransMultAdd(x, y);
+
+ double[] xd = ((DenseVector) x).getData(), yd = ((DenseVector) y)
+ .getData();
+
+ // y = 1/alpha * y
+ y.scale(1. / alpha);
+
+ // y = A'x + y
+ for (int i = 0; i < numRows; ++i) {
+ SparseVector v = rowD[i];
+ int[] index = v.getIndex();
+ double[] data = v.getData();
+ int length = v.getUsed();
+ for (int j = 0; j < length; ++j)
+ yd[index[j]] += data[j] * xd[i];
+ }
+
+ // y = alpha*y = alpha * A'x + y
+ return y.scale(alpha);
+ }
+
+ @Override
+ public void add(int row, int column, double value) {
+ rowD[row].add(column, value);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ rowD[row].set(column, value);
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return rowD[row].get(column);
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new RowMatrixIterator();
+ }
+
+ @Override
+ public Matrix copy() {
+ return new FlexCompRowMatrix(this);
+ }
+
+ @Override
+ public FlexCompRowMatrix zero() {
+ for (int i = 0; i < numRows; ++i)
+ rowD[i].zero();
+ return this;
+ }
+
+ @Override
+ public Matrix set(Matrix B) {
+ if (!(B instanceof FlexCompRowMatrix))
+ return super.set(B);
+
+ checkSize(B);
+
+ FlexCompRowMatrix Bc = (FlexCompRowMatrix) B;
+
+ for (int i = 0; i < numRows; ++i)
+ rowD[i].set(Bc.rowD[i]);
+
+ return this;
+ }
+
+ /**
+ * Tries to store the matrix as compactly as possible
+ */
+ public void compact() {
+ for (Vector v : rowD)
+ ((SparseVector) v).compact();
+ }
+
+ /**
+ * Iterator over a matrix stored vectorwise by rows
+ */
+ private class RowMatrixIterator implements Iterator<MatrixEntry> {
+
+ /**
+ * Iterates over each row vector
+ */
+ private SuperIterator<SparseVector, VectorEntry> iterator = new SuperIterator<SparseVector, VectorEntry>(
+ Arrays.asList(rowD));
+
+ /**
+ * Entry returned
+ */
+ private RowMatrixEntry entry = new RowMatrixEntry();
+
+ public boolean hasNext() {
+ return iterator.hasNext();
+ }
+
+ public MatrixEntry next() {
+ SuperIteratorEntry<VectorEntry> se = iterator.next();
+ entry.update(se.index(), se.get());
+ return entry;
+ }
+
+ public void remove() {
+ iterator.remove();
+ }
+
+ }
+
+ /**
+ * Entry of a matrix stored vectorwise by rows
+ */
+ private static class RowMatrixEntry implements MatrixEntry {
+
+ private int row;
+
+ private VectorEntry entry;
+
+ public void update(int row, VectorEntry entry) {
+ this.row = row;
+ this.entry = entry;
+ }
+
+ public int row() {
+ return row;
+ }
+
+ public int column() {
+ return entry.index();
+ }
+
+ public double get() {
+ return entry.get();
+ }
+
+ public void set(double value) {
+ entry.set(value);
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/GMRES.java b/src/main/java/no/uib/cipr/matrix/sparse/GMRES.java
new file mode 100644
index 0000000..e288f60
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/GMRES.java
@@ -0,0 +1,178 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.GivensRotation;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.UpperTriangDenseMatrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Vector.Norm;
+
+/**
+ * GMRES solver. GMRES solves the unsymmetric linear system <code>Ax = b</code>
+ * using the Generalized Minimum Residual method. The GMRES iteration is
+ * restarted after a given number of iterations. By default it is restarted
+ * after 30 iterations.
+ *
+ * @author Templates
+ */
+public class GMRES extends AbstractIterativeSolver {
+
+ /**
+ * After this many iterations, the GMRES will be restarted.
+ */
+ private int restart;
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector w, u, r;
+
+ /**
+ * Vectors spanning the subspace
+ */
+ private Vector[] v;
+
+ /**
+ * Restart vector
+ */
+ private DenseVector s;
+
+ /**
+ * Hessenberg matrix
+ */
+ private DenseMatrix H;
+
+ /**
+ * Givens rotations for the QR factorization
+ */
+ private GivensRotation[] rotation;
+
+ /**
+ * Constructor for GMRES. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified. The iteration is
+ * restarted every 30 iterations
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public GMRES(Vector template) {
+ this(template, 30);
+ }
+
+ /**
+ * Constructor for GMRES. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ * @param restart
+ * GMRES iteration is restarted after this number of iterations
+ */
+ public GMRES(Vector template, int restart) {
+ w = template.copy();
+ u = template.copy();
+ r = template.copy();
+ setRestart(restart);
+ }
+
+ /**
+ * Sets the restart parameter
+ *
+ * @param restart
+ * GMRES iteration is restarted after this number of iterations
+ */
+ public void setRestart(int restart) {
+ this.restart = restart;
+ if (restart <= 0)
+ throw new IllegalArgumentException(
+ "restart must be a positive integer");
+
+ s = new DenseVector(restart + 1);
+ H = new DenseMatrix(restart + 1, restart);
+ rotation = new GivensRotation[restart + 1];
+
+ v = new Vector[restart + 1];
+ for (int i = 0; i < v.length; ++i)
+ v[i] = r.copy().zero();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ A.multAdd(-1, x, u.set(b));
+ M.apply(u, r);
+ double normr = r.norm(Norm.Two);
+ M.apply(b, u);
+
+ // Outer iteration
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+
+ v[0].set(1 / normr, r);
+ s.zero().set(0, normr);
+ int i = 0;
+
+ // Inner iteration
+ for (; i < restart && !iter.converged(Math.abs(s.get(i))); i++, iter
+ .next()) {
+ A.mult(v[i], u);
+ M.apply(u, w);
+
+ for (int k = 0; k <= i; k++) {
+ H.set(k, i, w.dot(v[k]));
+ w.add(-H.get(k, i), v[k]);
+ }
+ H.set(i + 1, i, w.norm(Norm.Two));
+ v[i + 1].set(1. / H.get(i + 1, i), w);
+
+ // QR factorization of H using Givens rotations
+ for (int k = 0; k < i; ++k)
+ rotation[k].apply(H, i, k, k + 1);
+
+ rotation[i] = new GivensRotation(H.get(i, i), H.get(i + 1, i));
+ rotation[i].apply(H, i, i, i + 1);
+ rotation[i].apply(s, i, i + 1);
+ }
+
+ // Update solution in current subspace
+ new UpperTriangDenseMatrix(H, i, false).solve(s, s);
+ for (int j = 0; j < i; j++)
+ x.add(s.get(j), v[j]);
+
+ A.multAdd(-1, x, u.set(b));
+ M.apply(u, r);
+ normr = r.norm(Norm.Two);
+ }
+
+ return x;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/ICC.java b/src/main/java/no/uib/cipr/matrix/sparse/ICC.java
new file mode 100644
index 0000000..6192305
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/ICC.java
@@ -0,0 +1,155 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Arrays;
+
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Incomplete Cholesky preconditioner without fill-in using a compressed row
+ * matrix as internal storage
+ */
+public class ICC implements Preconditioner {
+
+ /**
+ * Factorisation matrix
+ */
+ private final CompRowMatrix R;
+
+ /**
+ * Triangular view onto R for solution purposes
+ */
+ private Matrix Rt;
+
+ /**
+ * Temporary vector for solving the factorised system
+ */
+ private final Vector y;
+
+ /**
+ * Sets up the ICC preconditioner
+ *
+ * @param R
+ * Matrix to use internally. For best performance, its non-zero
+ * pattern must conform to that of the system matrix
+ */
+ public ICC(CompRowMatrix R) {
+ if (!R.isSquare())
+ throw new IllegalArgumentException(
+ "ICC only applies to square matrices");
+
+ this.R = R;
+ int n = R.numRows();
+ y = new DenseVector(n);
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ // R'y = b, y = R'\b
+ Rt.transSolve(b, y);
+
+ // Rx = R'\b = y
+ return Rt.solve(y, x);
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ return apply(b, x);
+ }
+
+ public void setMatrix(Matrix A) {
+ R.set(A);
+
+ factor();
+ }
+
+ private void factor() {
+ int n = R.numRows();
+
+ // Internal CRS matrix storage
+ int[] colind = R.getColumnIndices();
+ int[] rowptr = R.getRowPointers();
+ double[] data = R.getData();
+
+ // Temporary storage of a dense row
+ double[] Rk = new double[n];
+
+ // Find the indices to the diagonal entries
+ int[] diagind = findDiagonalIndices(n, colind, rowptr);
+
+ // Go down along the main diagonal
+ for (int k = 0; k < n; ++k) {
+
+ // Expand current row to dense storage
+ Arrays.fill(Rk, 0);
+ for (int i = rowptr[k]; i < rowptr[k + 1]; ++i)
+ Rk[colind[i]] = data[i];
+
+ for (int i = 0; i < k; ++i) {
+
+ // Get the current diagonal entry
+ double Rii = data[diagind[i]];
+
+ if (Rii == 0)
+ throw new RuntimeException("Zero pivot encountered on row "
+ + (i + 1) + " during ICC process");
+
+ // Elimination factor
+ double Rki = Rk[i] / Rii;
+
+ if (Rki == 0)
+ continue;
+
+ // Traverse the sparse row i, reducing on row k
+ for (int j = diagind[i] + 1; j < rowptr[i + 1]; ++j)
+ Rk[colind[j]] -= Rki * data[j];
+ }
+
+ // Store the row back into the factorisation matrix
+ if (Rk[k] == 0)
+ throw new RuntimeException(
+ "Zero diagonal entry encountered on row " + (k + 1)
+ + " during ICC process");
+ double sqRkk = Math.sqrt(Rk[k]);
+
+ for (int i = diagind[k]; i < rowptr[k + 1]; ++i)
+ data[i] = Rk[colind[i]] / sqRkk;
+ }
+
+ Rt = new UpperCompRowMatrix(R, diagind);
+ }
+
+ private int[] findDiagonalIndices(int m, int[] colind, int[] rowptr) {
+ int[] diagind = new int[m];
+
+ for (int k = 0; k < m; ++k) {
+ diagind[k] = no.uib.cipr.matrix.sparse.Arrays.binarySearch(colind,
+ k, rowptr[k], rowptr[k + 1]);
+
+ if (diagind[k] < 0)
+ throw new RuntimeException("Missing diagonal entry on row "
+ + (k + 1));
+ }
+
+ return diagind;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/ILU.java b/src/main/java/no/uib/cipr/matrix/sparse/ILU.java
new file mode 100644
index 0000000..0a77103
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/ILU.java
@@ -0,0 +1,141 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * ILU(0) preconditioner using a compressed row matrix as internal storage
+ */
+public class ILU implements Preconditioner {
+
+ /**
+ * Factorisation matrix
+ */
+ private final CompRowMatrix LU;
+
+ /**
+ * The L and U factors
+ */
+ private Matrix L, U;
+
+ /**
+ * Temporary vector for solving the factorised system
+ */
+ private final Vector y;
+
+ /**
+ * Sets up the ILU preconditioner
+ *
+ * @param LU
+ * Matrix to use internally. For best performance, its non-zero
+ * pattern must conform to that of the system matrix
+ */
+ public ILU(CompRowMatrix LU) {
+ if (!LU.isSquare())
+ throw new IllegalArgumentException(
+ "ILU only applies to square matrices");
+
+ this.LU = LU;
+ int n = LU.numRows();
+ y = new DenseVector(n);
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ // Ly = b, y = L\b
+ L.solve(b, y);
+
+ // Ux = L\b = y
+ return U.solve(y, x);
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ // U'y = b, y = U'\b
+ U.transSolve(b, y);
+
+ // L'x = U'\b = y
+ return L.transSolve(y, x);
+ }
+
+ public void setMatrix(Matrix A) {
+ LU.set(A);
+
+ factor();
+ }
+
+ private void factor() {
+ int n = LU.numRows();
+
+ // Internal CRS matrix storage
+ int[] colind = LU.getColumnIndices();
+ int[] rowptr = LU.getRowPointers();
+ double[] data = LU.getData();
+
+ // Find the indices to the diagonal entries
+ int[] diagind = findDiagonalIndices(n, colind, rowptr);
+
+ // Go down along the main diagonal
+ for (int k = 1; k < n; ++k)
+ for (int i = rowptr[k]; i < diagind[k]; ++i) {
+
+ // Get the current diagonal entry
+ int index = colind[i];
+ double LUii = data[diagind[index]];
+
+ if (LUii == 0)
+ throw new RuntimeException("Zero pivot encountered on row "
+ + (i + 1) + " during ILU process");
+
+ // Elimination factor
+ double LUki = (data[i] /= LUii);
+
+ // Traverse the sparse row i, reducing on row k
+ for (int j = diagind[index] + 1, l = rowptr[k] + 1; j < rowptr[index + 1]; ++j) {
+
+ while (l < rowptr[k + 1] && colind[l] < colind[j])
+ l++;
+
+ if (colind[l] == colind[j])
+ data[l] -= LUki * data[j];
+ }
+ }
+
+ L = new UnitLowerCompRowMatrix(LU, diagind);
+ U = new UpperCompRowMatrix(LU, diagind);
+ }
+
+ private int[] findDiagonalIndices(int m, int[] colind, int[] rowptr) {
+ int[] diagind = new int[m];
+
+ for (int k = 0; k < m; ++k) {
+ diagind[k] = Arrays.binarySearch(colind, k, rowptr[k],
+ rowptr[k + 1]);
+
+ if (diagind[k] < 0)
+ throw new RuntimeException("Missing diagonal entry on row "
+ + (k + 1));
+ }
+
+ return diagind;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/ILUT.java b/src/main/java/no/uib/cipr/matrix/sparse/ILUT.java
new file mode 100644
index 0000000..37d4d3d
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/ILUT.java
@@ -0,0 +1,446 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.ArrayList;
+import java.util.Arrays;
+import java.util.Collections;
+import java.util.List;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.VectorEntry;
+
+/**
+ * ILU preconditioner with fill-in. Uses the dual threshold approach of Saad.
+ */
+public class ILUT implements Preconditioner {
+
+ /**
+ * Factorisation matrix
+ */
+ private final FlexCompRowMatrix LU;
+
+ /**
+ * The L and U factors
+ */
+ private Matrix L, U;
+
+ /**
+ * Temporary vector for solving the factorised system
+ */
+ private final Vector y;
+
+ /**
+ * Drop-tolerance
+ */
+ private final double tau;
+
+ /**
+ * Diagonal indices
+ */
+ private final int[] diagind;
+
+ /**
+ * Stores entries in the lower and upper part of the matrix. Used by the
+ * dropping rule to determine the largest entries in the two parts of the
+ * matrix
+ */
+ private final List<IntDoubleEntry> lower, upper;
+
+ /**
+ * Number of additional entries to keep in the lower and upper part of the
+ * factored matrix. The entries of the original matrix are always kept,
+ * unless they numerically too small
+ */
+ private final int p;
+
+ /**
+ * Sets up the preconditioner for the given matrix
+ *
+ * @param LU
+ * Matrix to use internally. For best performance, its non-zero
+ * pattern should conform to that of the system matrix
+ * @param tau
+ * Drop tolerance
+ * @param p
+ * Number of entries to keep on each row in of the factored
+ * matrix. This is in addition to the entries of the original
+ * matrix
+ */
+ public ILUT(FlexCompRowMatrix LU, double tau, int p) {
+ if (!LU.isSquare())
+ throw new IllegalArgumentException(
+ "ILU only applies to square matrices");
+
+ this.LU = LU;
+ this.tau = tau;
+ this.p = p;
+
+ int n = LU.numRows();
+ lower = new ArrayList<IntDoubleEntry>(n);
+ upper = new ArrayList<IntDoubleEntry>(n);
+ y = new DenseVector(n);
+ diagind = new int[n];
+ }
+
+ /**
+ * Sets up the preconditioner for the given matrix. Uses a drop-tolerance of
+ * 10<sup>-6</sup>, and keeps 50 entries on each row, including the main
+ * diagonal and any previous entries in the matrix structure
+ *
+ * @param LU
+ * Matrix to use internally. For best performance, its non-zero
+ * pattern should conform to that of the system matrix
+ */
+ public ILUT(FlexCompRowMatrix LU) {
+ this(LU, 1e-6, 25);
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ // Ly = b, y = L\b
+ L.solve(b, y);
+
+ // Ux = L\b = y
+ return U.solve(y, x);
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ // U'y = b, y = U'\b
+ U.transSolve(b, y);
+
+ // L'x = U'\b = y
+ return L.transSolve(y, x);
+ }
+
+ public void setMatrix(Matrix A) {
+ LU.set(A);
+ LU.compact();
+
+ factor();
+ }
+
+ private void factor() {
+ int n = LU.numRows();
+
+ double[] LUi = new double[n];
+
+ // Find the indices to the diagonal entries
+ for (int k = 0; k < n; ++k) {
+ SparseVector row = LU.getRow(k);
+ diagind[k] = findDiagonalIndex(row, k);
+ if (diagind[k] < 0)
+ throw new RuntimeException("Missing diagonal entry on row "
+ + (k + 1));
+ }
+
+ for (int i = 1; i < n; ++i) {
+
+ // Get row i
+ SparseVector rowi = LU.getRow(i);
+
+ // Drop tolerance on current row
+ double taui = rowi.norm(Vector.Norm.Two) * tau;
+
+ // Store in dense format
+ scatter(rowi, LUi);
+
+ for (int k = 0; k < i; ++k) {
+
+ // Get row k
+ SparseVector rowk = LU.getRow(k);
+ int[] rowIndex = rowk.getIndex();
+ int rowUsed = rowk.getUsed();
+ double[] rowData = rowk.getData();
+
+ if (rowData[diagind[k]] == 0)
+ throw new RuntimeException("Zero diagonal entry on row "
+ + (k + 1) + " during ILU process");
+
+ double LUik = LUi[k] / rowData[diagind[k]];
+
+ // Check for small elimination entry
+ if (Math.abs(LUik) <= taui)
+ continue;
+
+ // Traverse the sparse row k, reducing row i
+ for (int j = diagind[k] + 1; j < rowUsed; ++j)
+ LUi[rowIndex[j]] -= LUik * rowData[j];
+
+ // The above has overwritten LUik, so remedy that
+ LUi[k] = LUik;
+ }
+
+ // Store back into the LU matrix, dropping as needed
+ gather(LUi, rowi, taui, i);
+
+ // Update diagonal index on row i if it is outdated
+ if (rowi.getIndex()[diagind[i]] != i) {
+ diagind[i] = findDiagonalIndex(rowi, i);
+ if (diagind[i] < 0)
+ throw new RuntimeException("Missing diagonal entry on row "
+ + (i + 1) + " during ILU process");
+ }
+ }
+
+ L = new UnitLowerFlexCompRowMatrix(LU, diagind);
+ U = new UpperFlexCompRowMatrix(LU, diagind);
+ }
+
+ private int findDiagonalIndex(SparseVector v, int k) {
+ return no.uib.cipr.matrix.sparse.Arrays.binarySearch(v.getIndex(), k,
+ 0, v.getUsed());
+ }
+
+ /**
+ * Copies the sparse vector into a dense array
+ */
+ private void scatter(SparseVector v, double[] z) {
+ int[] index = v.getIndex();
+ int used = v.getUsed();
+ double[] data = v.getData();
+ Arrays.fill(z, 0);
+ for (int i = 0; i < used; ++i)
+ z[index[i]] = data[i];
+ }
+
+ /**
+ * Copies the dense array back into the sparse vector, applying a numerical
+ * dropping rule and keeping only a given number of entries
+ */
+ private void gather(double[] z, SparseVector v, double taui, int d) {
+ // Number of entries in the lower and upper part of the original matrix
+ int nl = 0, nu = 0;
+ for (VectorEntry e : v) {
+ if (e.index() < d)
+ nl++;
+ else if (e.index() > d)
+ nu++;
+ }
+ v.zero();
+
+ // Entries in the L part of the vector
+ lower.clear();
+ for (int i = 0; i < d; ++i)
+ if (Math.abs(z[i]) > taui)
+ lower.add(new IntDoubleEntry(i, z[i]));
+
+ // Entries in the U part of the vector
+ upper.clear();
+ for (int i = d + 1; i < z.length; ++i)
+ if (Math.abs(z[i]) > taui)
+ upper.add(new IntDoubleEntry(i, z[i]));
+
+ // Sort in descending order
+ Collections.sort(lower);
+ Collections.sort(upper);
+
+ // Always keep the diagonal
+ v.set(d, z[d]);
+
+ // Keep at most nl+p lower entries
+ for (int i = 0; i < Math.min(nl + p, lower.size()); ++i) {
+ IntDoubleEntry e = lower.get(i);
+ v.set(e.index, e.value);
+ }
+
+ // Keep at most nu+p upper entries
+ for (int i = 0; i < Math.min(nu + p, upper.size()); ++i) {
+ IntDoubleEntry e = upper.get(i);
+ v.set(e.index, e.value);
+ }
+ }
+
+ /**
+ * Stores an integer/value pair, sorted by descending order according to the
+ * value
+ */
+ private static class IntDoubleEntry implements Comparable<IntDoubleEntry> {
+
+ public int index;
+
+ public double value;
+
+ public IntDoubleEntry(int index, double value) {
+ this.index = index;
+ this.value = value;
+ }
+
+ public int compareTo(IntDoubleEntry o) {
+ // Descending order, so keep the largest entries first
+ if (Math.abs(value) < Math.abs(o.value))
+ return 1;
+ else if (Math.abs(value) == Math.abs(o.value))
+ return 0;
+ else
+ return -1;
+ }
+
+ @Override
+ public String toString() {
+ return "(" + index + "=" + value + ")";
+ }
+ }
+
+ /**
+ * Unit lower triangular flex-CRS matrix. Only used for triangular solves
+ */
+ private static class UnitLowerFlexCompRowMatrix extends AbstractMatrix {
+
+ private final FlexCompRowMatrix LU;
+
+ private final int[] diagind;
+
+ public UnitLowerFlexCompRowMatrix(FlexCompRowMatrix LU, int[] diagind) {
+ super(LU);
+ this.LU = LU;
+ this.diagind = diagind;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ if (!(b instanceof DenseVector) || !(x instanceof DenseVector))
+ return super.solve(b, x);
+
+ double[] bd = ((DenseVector) b).getData();
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = 0; i < numRows; ++i) {
+
+ // Get row i
+ SparseVector row = LU.getRow(i);
+ int[] index = row.getIndex();
+ double[] data = row.getData();
+
+ // xi = bi - sum[j<i] Lij * xj
+ double sum = 0;
+ for (int j = 0; j < diagind[i]; ++j)
+ sum += data[j] * xd[index[j]];
+
+ xd[i] = bd[i] - sum;
+ }
+
+ return x;
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ if (!(x instanceof DenseVector))
+ return super.transSolve(b, x);
+
+ x.set(b);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = numRows - 1; i >= 0; --i) {
+
+ // Get row i
+ SparseVector row = LU.getRow(i);
+ int[] index = row.getIndex();
+ double[] data = row.getData();
+
+ // At this stage, x[i] is known, so move it over to the right
+ // hand side for the remaining equations
+ for (int j = 0; j < diagind[i]; ++j)
+ xd[index[j]] -= data[j] * xd[i];
+
+ }
+
+ return x;
+ }
+
+ }
+
+ /**
+ * Upper triangular flex-CRS matrix. Only used for triangular solves
+ */
+ private static class UpperFlexCompRowMatrix extends AbstractMatrix {
+
+ private final FlexCompRowMatrix LU;
+
+ private final int[] diagind;
+
+ public UpperFlexCompRowMatrix(FlexCompRowMatrix LU, int[] diagind) {
+ super(LU);
+ this.LU = LU;
+ this.diagind = diagind;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ if (!(b instanceof DenseVector) || !(x instanceof DenseVector))
+ return super.solve(b, x);
+
+ double[] bd = ((DenseVector) b).getData();
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = numRows - 1; i >= 0; --i) {
+
+ // Get row i
+ SparseVector row = LU.getRow(i);
+ int[] index = row.getIndex();
+ int used = row.getUsed();
+ double[] data = row.getData();
+
+ // xi = (bi - sum[j>i] Uij * xj) / Uii
+ double sum = 0;
+ for (int j = diagind[i] + 1; j < used; ++j)
+ sum += data[j] * xd[index[j]];
+
+ xd[i] = (bd[i] - sum) / data[diagind[i]];
+ }
+
+ return x;
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ if (!(x instanceof DenseVector))
+ return super.transSolve(b, x);
+
+ x.set(b);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = 0; i < numRows; ++i) {
+
+ // Get row i
+ SparseVector row = LU.getRow(i);
+ int[] index = row.getIndex();
+ int used = row.getUsed();
+ double[] data = row.getData();
+
+ // Solve for the current entry
+ xd[i] /= data[diagind[i]];
+
+ // Move this known solution over to the right hand side for the
+ // remaining equations
+ for (int j = diagind[i] + 1; j < used; ++j)
+ xd[index[j]] -= data[j] * xd[i];
+ }
+
+ return x;
+ }
+
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/IR.java b/src/main/java/no/uib/cipr/matrix/sparse/IR.java
new file mode 100644
index 0000000..06dc031
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/IR.java
@@ -0,0 +1,72 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Iterative Refinement. IR solves the unsymmetric linear system
+ * <code>Ax = b</code> using Iterative Refinement (preconditioned Richardson
+ * iteration).
+ *
+ * @author Templates
+ */
+public class IR extends AbstractIterativeSolver {
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector z, r;
+
+ /**
+ * Constructor for IR. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public IR(Vector template) {
+ z = template.copy();
+ r = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ A.multAdd(-1, x, r.set(b));
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+ M.apply(r, z);
+ x.add(z);
+ A.multAdd(-1, x, r.set(b));
+ }
+
+ return x;
+ }
+
+}
\ No newline at end of file
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/ISparseVector.java b/src/main/java/no/uib/cipr/matrix/sparse/ISparseVector.java
new file mode 100644
index 0000000..a5d9a56
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/ISparseVector.java
@@ -0,0 +1,24 @@
+/**
+ * May 21, 2007
+ * @author Samuel Halliday, ThinkTank Maths Limited
+ * Copyright ThinkTank Maths Limited 2007
+ */
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * @author Samuel Halliday, ThinkTank Maths Limited
+ */
+public interface ISparseVector extends Vector {
+
+ /**
+ * Returns the indices
+ */
+ public int[] getIndex();
+
+ /**
+ * Number of entries used in the sparse structure
+ */
+ public int getUsed();
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/IterationMonitor.java b/src/main/java/no/uib/cipr/matrix/sparse/IterationMonitor.java
new file mode 100644
index 0000000..dc16fa3
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/IterationMonitor.java
@@ -0,0 +1,119 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Vector.Norm;
+
+/**
+ * Monitors the iterative solution process for convergence and divergence. Can
+ * also report the current progress.
+ */
+public interface IterationMonitor {
+
+ /**
+ * Resets the iteration
+ */
+ void setFirst();
+
+ /**
+ * Returns true for the first iteration
+ */
+ boolean isFirst();
+
+ /**
+ * Increases iteration counter
+ */
+ void next();
+
+ /**
+ * Number of iterations performed
+ */
+ int iterations();
+
+ /**
+ * Returns current residual
+ */
+ double residual();
+
+ /**
+ * Checks for convergence
+ *
+ * @param r
+ * Residual-vector
+ * @param x
+ * State-vector
+ * @return True if converged
+ */
+ boolean converged(Vector r, Vector x)
+ throws IterativeSolverNotConvergedException;
+
+ /**
+ * Checks for convergence
+ *
+ * @param r
+ * Residual-norm
+ * @param x
+ * State-vector
+ * @return True if converged
+ */
+ boolean converged(double r, Vector x)
+ throws IterativeSolverNotConvergedException;
+
+ /**
+ * Checks for convergence
+ *
+ * @param r
+ * Residual-norm
+ * @return True if converged
+ */
+ boolean converged(double r) throws IterativeSolverNotConvergedException;
+
+ /**
+ * Checks for convergence
+ *
+ * @param r
+ * Residual-vector
+ * @return True if converged
+ */
+ boolean converged(Vector r) throws IterativeSolverNotConvergedException;
+
+ /**
+ * Sets new iteration reporter
+ */
+ void setIterationReporter(IterationReporter monitor);
+
+ /**
+ * Returns current iteration reporter
+ */
+ IterationReporter getIterationReporter();
+
+ /**
+ * Sets the vector-norm to calculate with
+ */
+ void setNormType(Norm normType);
+
+ /**
+ * Returns the vector-norm in use
+ */
+ Norm getNormType();
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/IterationReporter.java b/src/main/java/no/uib/cipr/matrix/sparse/IterationReporter.java
new file mode 100644
index 0000000..1a68d2b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/IterationReporter.java
@@ -0,0 +1,52 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Reports on the progress of an iterative solver
+ */
+public interface IterationReporter {
+
+ /**
+ * Registers current information
+ *
+ * @param r
+ * Current residual norm
+ * @param x
+ * Current state vector
+ * @param i
+ * Current iteration number
+ */
+ void monitor(double r, Vector x, int i);
+
+ /**
+ * Registers current information
+ *
+ * @param r
+ * Current residual norm
+ * @param i
+ * Current iteration number
+ */
+ void monitor(double r, int i);
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolver.java b/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolver.java
new file mode 100644
index 0000000..efde04c
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolver.java
@@ -0,0 +1,76 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Iterative linear solver. Solves <code>Ax=b</code> for <code>x</code>,
+ * and it supports preconditioning and convergence monitoring.
+ */
+public interface IterativeSolver {
+
+ /**
+ * Solves the given problem, writing result into the vector.
+ *
+ * @param A
+ * Matrix of the problem
+ * @param b
+ * Right hand side
+ * @param x
+ * Solution is stored here. Also used as initial guess
+ * @return The solution vector x
+ */
+ Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException;
+
+ /**
+ * Sets preconditioner
+ *
+ * @param M
+ * Preconditioner to use
+ */
+ void setPreconditioner(Preconditioner M);
+
+ /**
+ * Gets preconditioner
+ *
+ * @return Current preconditioner
+ */
+ Preconditioner getPreconditioner();
+
+ /**
+ * Sets iteration monitor
+ *
+ * @param iter
+ * Iteration monitor
+ */
+ void setIterationMonitor(IterationMonitor iter);
+
+ /**
+ * Gets the iteration monitor
+ *
+ * @return Current iteration monitor
+ */
+ IterationMonitor getIterationMonitor();
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolverNotConvergedException.java b/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolverNotConvergedException.java
new file mode 100644
index 0000000..51f2647
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/IterativeSolverNotConvergedException.java
@@ -0,0 +1,91 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.NotConvergedException;
+
+/**
+ * Exception for lack of convergence in a linear problem. Contains the final
+ * computed residual.
+ */
+public class IterativeSolverNotConvergedException extends NotConvergedException {
+
+ private static final long serialVersionUID = 5354102050137093202L;
+
+ /**
+ * Iteration count when this exception was thrown
+ */
+ private int iterations;
+
+ /**
+ * Final residual
+ */
+ private double r;
+
+ /**
+ * Constructor for IterativeSolverNotConvergedException
+ *
+ * @param reason
+ * Reason for this exception
+ * @param message
+ * A more detailed message
+ * @param iter
+ * Associated iteration monitor, for extracting residual and
+ * iteration number
+ */
+ public IterativeSolverNotConvergedException(Reason reason, String message,
+ IterationMonitor iter) {
+ super(reason, message);
+ this.r = iter.residual();
+ this.iterations = iter.iterations();
+ }
+
+ /**
+ * Constructor for IterativeSolverNotConvergedException
+ *
+ * @param reason
+ * Reason for this exception
+ * @param iter
+ * Associated iteration monitor, for extracting residual and
+ * iteration number
+ */
+ public IterativeSolverNotConvergedException(Reason reason,
+ IterationMonitor iter) {
+ super(reason);
+ this.r = iter.residual();
+ this.iterations = iter.iterations();
+ }
+
+ /**
+ * Returns final computed residual
+ */
+ public double getResidual() {
+ return r;
+ }
+
+ /**
+ * Gets the number of iterations used when this exception was thrown
+ */
+ public int getIterations() {
+ return iterations;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrix.java
new file mode 100644
index 0000000..04d9aaf
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrix.java
@@ -0,0 +1,457 @@
+package no.uib.cipr.matrix.sparse;
+
+import lombok.AllArgsConstructor;
+import lombok.ToString;
+import lombok.extern.java.Log;
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.io.MatrixInfo;
+import no.uib.cipr.matrix.io.MatrixSize;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+import java.io.IOException;
+import java.util.Iterator;
+
+/**
+ * A Linked List (with shortcuts to important nodes)
+ * implementation of an {@code n x m} Matrix with {@code z} elements that
+ * has a typical {@code O(z / m)} insertion / lookup cost
+ * and an iterator that traverses columns then rows:
+ * a good fit for unstructured sparse matrices. A secondary
+ * link maintains fast transpose iteration.
+ * <p/>
+ * However, memory requirements
+ * ({@code 1 instance (8 bytes), 2 int (16 bytes), 2 ref (16 bytes), 1 double (8 bytes) = 48 bytes}
+ * per matrix element, plus {@code 8 x numcol + 8 x numrow bytes}s for the cache) are slightly higher
+ * than structured sparse matrix storage. Note that on 32 bit JVMs, or on 64 bit JVMs
+ * with <a href="https://wikis.oracle.com/display/HotSpotInternals/CompressedOops">CompressedOops</a>
+ * enabled, references and ints only cost 4 bytes each, bringing the cost to 28 bytes per element.
+ *
+ * @author Sam Halliday
+ */
+ at Log
+public class LinkedSparseMatrix extends AbstractMatrix {
+
+ // java.util.LinkedList is doubly linked and therefore too heavyweight.
+ @AllArgsConstructor
+ @ToString(exclude = {"rowTail", "colTail"})
+ static class Node {
+ final int row, col;
+ double val;
+ Node rowTail, colTail;
+ }
+
+ // there is a lot of duplicated code in this class between
+ // row and col linkages, but subtle differences make it
+ // extremely difficult to factor away.
+ class Linked {
+
+ final Node head = new Node(0, 0, 0, null, null);
+
+ Node[] rows = new Node[numRows], cols = new Node[numColumns];
+
+ private boolean isHead(int row, int col) {
+ return head.row == row && head.col == col;
+ }
+
+ // true if node exists, it's row tail exists, and has this row/col
+ private boolean isNextByRow(Node node, int row, int col) {
+ return node != null && node.rowTail != null && node.rowTail.row == row && node.rowTail.col == col;
+ }
+
+ // true if node exists, it's col tail exists, and has this row/col
+ private boolean isNextByCol(Node node, int row, int col) {
+ return node != null && node.colTail != null && node.colTail.col == col && node.colTail.row == row;
+ }
+
+ public double get(int row, int col) {
+ if (isHead(row, col))
+ return head.val;
+ if (col <= row) {
+ Node node = findPreceedingByRow(row, col);
+ if (isNextByRow(node, row, col))
+ return node.rowTail.val;
+ } else {
+ Node node = findPreceedingByCol(row, col);
+ if (isNextByCol(node, row, col))
+ return node.colTail.val;
+ }
+ return 0;
+ }
+
+ public void set(int row, int col, double val) {
+ if (val == 0) {
+ delete(row, col);
+ return;
+ }
+ if (isHead(row, col))
+ head.val = val;
+ else {
+ Node prevRow = findPreceedingByRow(row, col);
+ if (isNextByRow(prevRow, row, col))
+ prevRow.rowTail.val = val;
+ else {
+ Node prevCol = findPreceedingByCol(row, col);
+ Node nextCol = findNextByCol(row, col);
+ prevRow.rowTail = new Node(row, col, val, prevRow.rowTail, nextCol);
+ prevCol.colTail = prevRow.rowTail;
+ updateCache(prevRow.rowTail);
+ }
+ }
+ // DEBUGGING
+ if (isHead(row, col))
+ assert head.val == val;
+ else {
+ Node node = findPreceedingByCol(row, col);
+ assert node != null;
+ assert node.colTail.val == val;
+ }
+ }
+
+ private Node findNextByCol(int row, int col) {
+ Node cur = cachedByCol(col - 1);
+ while (cur != null) {
+ if (row < cur.row && col <= cur.col || col < cur.col) return cur;
+ cur = cur.colTail;
+ }
+ return cur;
+ }
+
+ private void updateCache(Node inserted) {
+ if (rows[inserted.row] == null || inserted.col > rows[inserted.row].col)
+ rows[inserted.row] = inserted;
+ if (cols[inserted.col] == null || inserted.row > cols[inserted.col].row)
+ cols[inserted.col] = inserted;
+ }
+
+ private void delete(int row, int col) {
+ if (isHead(row, col)) {
+ head.val = 0;
+ return;
+ }
+ Node precRow = findPreceedingByRow(row, col);
+ Node precCol = findPreceedingByCol(row, col);
+ if (isNextByRow(precRow, row, col)) {
+ if (rows[row] == precRow.rowTail)
+ rows[row] = precRow.row == row ? precRow : null;
+ precRow.rowTail = precRow.rowTail.rowTail;
+ }
+ if (isNextByCol(precCol, row, col)) {
+ if (cols[col] == precCol.colTail)
+ cols[col] = precCol.col == col ? precCol : null;
+ precCol.colTail = precCol.colTail.colTail;
+ }
+ }
+
+ // returns the node that either references this
+ // index, or should reference it if inserted.
+ Node findPreceedingByRow(int row, int col) {
+ Node last = cachedByRow(row - 1);
+ Node cur = last;
+ while (cur != null && cur.row <= row) {
+ if (cur.row == row && cur.col >= col) return last;
+ last = cur;
+ cur = cur.rowTail;
+ }
+ return last;
+ }
+
+ // helper for findPreceeding
+ private Node cachedByRow(int row) {
+ for (int i = row; i >= 0; i--)
+ if (rows[i] != null)
+ return rows[i];
+ return head;
+ }
+
+ Node findPreceedingByCol(int row, int col) {
+ Node last = cachedByCol(col - 1);
+ Node cur = last;
+ while (cur != null && cur.col <= col) {
+ if (cur.col == col && cur.row >= row) return last;
+ last = cur;
+ cur = cur.colTail;
+ }
+ return last;
+ }
+
+ private Node cachedByCol(int col) {
+ for (int i = col; i >= 0; i--)
+ if (cols[i] != null)
+ return cols[i];
+ return head;
+ }
+
+ Node startOfRow(int row) {
+ if (row == 0) return head;
+ Node prec = findPreceedingByRow(row, 0);
+ if (prec.rowTail != null && prec.rowTail.row == row)
+ return prec.rowTail;
+ return null;
+ }
+
+ Node startOfCol(int col) {
+ if (col == 0) return head;
+ Node prec = findPreceedingByCol(0, col);
+ if (prec != null && prec.colTail != null && prec.colTail.col == col)
+ return prec.colTail;
+ return null;
+ }
+ }
+
+ Linked links;
+
+ protected LinkedSparseMatrix(int numRows, int numColumns) {
+ super(numRows, numColumns);
+ links = new Linked();
+ }
+
+ protected LinkedSparseMatrix(Matrix A) {
+ super(A);
+ links = new Linked();
+ set(A);
+ }
+
+ public LinkedSparseMatrix(MatrixVectorReader r) throws IOException {
+ super(0, 0);
+ try {
+ MatrixInfo info = r.readMatrixInfo();
+ if (info.isComplex()) throw new IllegalArgumentException("complex matrices not supported");
+ if (!info.isCoordinate()) throw new IllegalArgumentException("only coordinate matrices supported");
+ MatrixSize size = r.readMatrixSize(info);
+ numRows = size.numRows();
+ numColumns = size.numColumns();
+ links = new Linked();
+
+ int nz = size.numEntries();
+ int[] row = new int[nz];
+ int[] column = new int[nz];
+ double[] entry = new double[nz];
+ r.readCoordinate(row, column, entry);
+ r.add(-1, row);
+ r.add(-1, column);
+ for (int i = 0; i < nz; ++i)
+ set(row[i], column[i], entry[i]);
+ } finally {
+ r.close();
+ }
+ }
+
+
+ @Override
+ public Matrix zero() {
+ links = new Linked();
+ return this;
+ }
+
+ @Override
+ public double get(int row, int column) {
+ return links.get(row, column);
+ }
+
+ @Override
+ public void set(int row, int column, double value) {
+ check(row, column);
+ links.set(row, column, value);
+ }
+
+ // avoids object creation
+ static class ReusableMatrixEntry implements MatrixEntry {
+
+ int row, col;
+ double val;
+
+ @Override
+ public int column() {
+ return col;
+ }
+
+ @Override
+ public int row() {
+ return row;
+ }
+
+ @Override
+ public double get() {
+ return val;
+ }
+
+ @Override
+ public void set(double value) {
+ throw new UnsupportedOperationException();
+ }
+
+ @Override
+ public String toString() {
+ return row + "," + col + "=" + val;
+ }
+
+ }
+
+ @Override
+ public Iterator<MatrixEntry> iterator() {
+ return new Iterator<MatrixEntry>() {
+ Node cur = links.head;
+ ReusableMatrixEntry entry = new ReusableMatrixEntry();
+
+ @Override
+ public boolean hasNext() {
+ return cur != null;
+ }
+
+ @Override
+ public MatrixEntry next() {
+ entry.row = cur.row;
+ entry.col = cur.col;
+ entry.val = cur.val;
+ cur = cur.rowTail;
+ return entry;
+ }
+
+ @Override
+ public void remove() {
+ throw new UnsupportedOperationException("TODO");
+ }
+ };
+ }
+
+ @Override
+ public Matrix scale(double alpha) {
+ if (alpha == 0)
+ zero();
+ else if (alpha != 1) for (MatrixEntry e : this)
+ set(e.row(), e.column(), e.get() * alpha);
+ return this;
+ }
+
+ @Override
+ public Matrix copy() {
+ return new LinkedSparseMatrix(this);
+ }
+
+ @Override
+ public Matrix transpose() {
+ Linked old = links;
+ numRows = numColumns;
+ numColumns = old.rows.length;
+ links = new Linked();
+ Node node = old.head;
+ while (node != null) {
+ set(node.col, node.row, node.val);
+ node = node.rowTail;
+ }
+ return this;
+ }
+
+ @Override
+ public Vector multAdd(double alpha, Vector x, Vector y) {
+ checkMultAdd(x, y);
+ if (alpha == 0) return y;
+ Node node = links.head;
+ while (node != null) {
+ y.add(node.row, alpha * node.val * x.get(node.col));
+ node = node.rowTail;
+ }
+ return y;
+ }
+
+ @Override
+ public Vector transMultAdd(double alpha, Vector x, Vector y) {
+ checkTransMultAdd(x, y);
+ if (alpha == 0) return y;
+ Node node = links.head;
+ while (node != null) {
+ y.add(node.col, alpha * node.val * x.get(node.row));
+ node = node.colTail;
+ }
+ return y;
+ }
+
+ // TODO: optimise matrix mults based on RHS Matrix
+
+ @Override
+ public Matrix multAdd(double alpha, Matrix B, Matrix C) {
+ checkMultAdd(B, C);
+ if (alpha == 0)
+ return C;
+ for (int i = 0; i < numRows; i++) {
+ Node row = links.startOfRow(i);
+ if (row != null)
+ for (int j = 0; j < B.numColumns(); j++) {
+ Node node = row;
+ double v = 0;
+ while (node != null && node.row == i) {
+ v += (B.get(node.col, j) * node.val);
+ node = node.rowTail;
+ }
+ if (v != 0) C.add(i, j, alpha * v);
+ }
+ }
+ return C;
+ }
+
+ @Override
+ public Matrix transBmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransBmultAdd(B, C);
+ if (alpha == 0)
+ return C;
+ for (int i = 0; i < numRows; i++) {
+ Node row = links.startOfRow(i);
+ if (row != null)
+ for (int j = 0; j < B.numRows(); j++) {
+ Node node = row;
+ double v = 0;
+ while (node != null && node.row == i) {
+ v += (B.get(j, node.col) * node.val);
+ node = node.rowTail;
+ }
+ if (v != 0) C.add(i, j, alpha * v);
+ }
+ }
+ return C;
+ }
+
+ @Override
+ public Matrix transAmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransAmultAdd(B, C);
+ if (alpha == 0)
+ return C;
+ for (int i = 0; i < numColumns; i++) {
+ Node row = links.startOfCol(i);
+ if (row != null)
+ for (int j = 0; j < B.numColumns(); j++) {
+ Node node = row;
+ double v = 0;
+ while (node != null && node.col == i) {
+ v += (B.get(node.row, j) * node.val);
+ node = node.colTail;
+ }
+ if (v != 0) C.add(i, j, alpha * v);
+ }
+ }
+ return C;
+ }
+
+ @Override
+ public Matrix transABmultAdd(double alpha, Matrix B, Matrix C) {
+ checkTransABmultAdd(B, C);
+ if (alpha == 0)
+ return C;
+ for (int i = 0; i < numColumns; i++) {
+ Node row = links.startOfCol(i);
+ if (row != null)
+ for (int j = 0; j < B.numRows(); j++) {
+ Node node = row;
+ double v = 0;
+ while (node != null && node.col == i) {
+ v += (B.get(j, node.row) * node.val);
+ node = node.colTail;
+ }
+ if (v != 0) C.add(i, j, alpha * v);
+ }
+ }
+ return C;
+ }
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/MatrixIterationMonitor.java b/src/main/java/no/uib/cipr/matrix/sparse/MatrixIterationMonitor.java
new file mode 100644
index 0000000..1d00b89
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/MatrixIterationMonitor.java
@@ -0,0 +1,131 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Iteration monitor based on matrix norms. Extends the default linear iteration
+ * object to compare with the norm of the system matrix and the right hand side.
+ * Can often be a better convergence criteria than the default, but requires the
+ * computation of the matrix norm.
+ */
+public class MatrixIterationMonitor extends DefaultIterationMonitor {
+
+ /**
+ * Norm of the system matrix
+ */
+ private double normA;
+
+ /**
+ * Norm of the right hand side
+ */
+ private double normb;
+
+ /**
+ * Constructor for MatrixIterationMonitor
+ *
+ * @param normA
+ * Norm of the matrix A
+ * @param normb
+ * Norm of the vector b
+ * @param maxIter
+ * Maximum number of iterations
+ * @param rtol
+ * Relative convergence tolerance (to initial residual)
+ * @param atol
+ * Absolute convergence tolerance
+ * @param dtol
+ * Relative divergence tolerance (to initial residual)
+ */
+ public MatrixIterationMonitor(double normA, double normb, int maxIter,
+ double rtol, double atol, double dtol) {
+ this.normA = normA;
+ this.normb = normb;
+ this.maxIter = maxIter;
+ this.rtol = rtol;
+ this.atol = atol;
+ this.dtol = dtol;
+ }
+
+ /**
+ * Constructor for MatrixIterationMonitor. Default is 100000 iterations at
+ * most, relative tolerance of 1e-5, absolute tolerance of 1e-50 and a
+ * divergence tolerance of 1e+5.
+ */
+ public MatrixIterationMonitor(double normA, double normb) {
+ this.normA = normA;
+ this.normb = normb;
+ }
+
+ /**
+ * Sets the norm of the system matrix
+ *
+ * @param normA
+ * Norm of the matrix A
+ */
+ public void setMatrixNorm(double normA) {
+ this.normA = normA;
+ }
+
+ /**
+ * Sets the norm of the right hand side vector
+ *
+ * @param normb
+ * Norm of the vector b
+ */
+ public void setVectorNorm(double normb) {
+ this.normb = normb;
+ }
+
+ @Override
+ protected boolean convergedI(double r, Vector x)
+ throws IterativeSolverNotConvergedException {
+ // Store initial residual
+ if (isFirst())
+ initR = r;
+
+ // Check for convergence
+ if (r < Math.max(rtol * (normA * x.norm(normType) + normb), atol))
+ return true;
+
+ // Check for divergence
+ if (r > dtol * initR)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Divergence, this);
+ if (iter >= maxIter)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Iterations, this);
+ if (Double.isNaN(r))
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Divergence, this);
+
+ // Neither convergence nor divergence
+ return false;
+ }
+
+ @Override
+ protected boolean convergedI(double r) {
+ throw new UnsupportedOperationException();
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/NoIterationReporter.java b/src/main/java/no/uib/cipr/matrix/sparse/NoIterationReporter.java
new file mode 100644
index 0000000..46b7f61
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/NoIterationReporter.java
@@ -0,0 +1,38 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * An iteration reporter which does nothing.
+ */
+public class NoIterationReporter implements IterationReporter {
+
+ public void monitor(double r, int i) {
+ // void
+ }
+
+ public void monitor(double r, Vector x, int i) {
+ // void
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/OutputIterationReporter.java b/src/main/java/no/uib/cipr/matrix/sparse/OutputIterationReporter.java
new file mode 100644
index 0000000..031030d
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/OutputIterationReporter.java
@@ -0,0 +1,64 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.io.OutputStream;
+import java.io.PrintWriter;
+
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Outputs iteration information to an output stream.
+ */
+public class OutputIterationReporter implements IterationReporter {
+
+ /**
+ * Platform-dependent output
+ */
+ private PrintWriter out;
+
+ /**
+ * Constructor for OutputIterationReporter
+ *
+ * @param out
+ * Writes iteration count and current residual here
+ */
+ public OutputIterationReporter(OutputStream out) {
+ this.out = new PrintWriter(out, true);
+ }
+
+ /**
+ * Constructor for OutputIterationReporter, using <code>System.err</code>.
+ */
+ public OutputIterationReporter() {
+ this(System.err);
+ }
+
+ public void monitor(double r, int i) {
+ out.format("%10d % .12e\n", i, r);
+ out.flush();
+ }
+
+ public void monitor(double r, Vector x, int i) {
+ monitor(r, i);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/Preconditioner.java b/src/main/java/no/uib/cipr/matrix/sparse/Preconditioner.java
new file mode 100644
index 0000000..6167edf
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/Preconditioner.java
@@ -0,0 +1,65 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Preconditioner interface. Before a preconditioner is used,
+ * <code>setMatrix</code> must be called
+ */
+public interface Preconditioner {
+
+ /**
+ * Solves the approximate problem with the given right hand side. Result is
+ * stored in given solution vector
+ *
+ * @param b
+ * Right hand side of problem
+ * @param x
+ * Result is stored here
+ * @return x
+ */
+ Vector apply(Vector b, Vector x);
+
+ /**
+ * Solves the approximate transpose problem with the given right hand side.
+ * Result is stored in given solution vector
+ *
+ * @param b
+ * Right hand side of problem
+ * @param x
+ * Result is stored here
+ * @return x
+ */
+ Vector transApply(Vector b, Vector x);
+
+ /**
+ * Sets the operator matrix for the preconditioner. This method must be
+ * called before a preconditioner is used by an iterative solver
+ *
+ * @param A
+ * Matrix to setup the preconditioner for. Not modified
+ */
+ void setMatrix(Matrix A);
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/QMR.java b/src/main/java/no/uib/cipr/matrix/sparse/QMR.java
new file mode 100644
index 0000000..f7c2454
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/QMR.java
@@ -0,0 +1,222 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+/*
+ * Derived from public domain software at http://www.netlib.org/templates
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Vector.Norm;
+
+/**
+ * Quasi-Minimal Residual method. QMR solves the unsymmetric linear system
+ * <code>Ax = b</code> using the Quasi-Minimal Residual method. QMR uses two
+ * preconditioners, and by default these are the same preconditioner.
+ *
+ * @author Templates
+ */
+public class QMR extends AbstractIterativeSolver {
+
+ /**
+ * Left preconditioner
+ */
+ private Preconditioner M1;
+
+ /**
+ * Right preconditioner
+ */
+ private Preconditioner M2;
+
+ /**
+ * Vectors for use in the iterative solution process
+ */
+ private Vector r, y, z, v, w, p, q, d, s, v_tld, w_tld, y_tld, z_tld,
+ p_tld;
+
+ /**
+ * Constructor for QMR. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ */
+ public QMR(Vector template) {
+ M1 = M;
+ M2 = M;
+ r = template.copy();
+ y = template.copy();
+ z = template.copy();
+ v = template.copy();
+ w = template.copy();
+ p = template.copy();
+ q = template.copy();
+ d = template.copy();
+ s = template.copy();
+ v_tld = template.copy();
+ w_tld = template.copy();
+ y_tld = template.copy();
+ z_tld = template.copy();
+ p_tld = template.copy();
+ }
+
+ /**
+ * Constructor for QMR. Uses the given vector as template for creating
+ * scratch vectors. Typically, the solution or the right hand side vector
+ * can be passed, and the template is not modified. Allows setting different
+ * right and left preconditioners
+ *
+ * @param template
+ * Vector to use as template for the work vectors needed in the
+ * solution process
+ * @param M1
+ * Left preconditioner
+ * @param M2
+ * Right preconditioner
+ */
+ public QMR(Vector template, Preconditioner M1, Preconditioner M2) {
+ this.M1 = M1;
+ this.M2 = M2;
+ r = template.copy();
+ y = template.copy();
+ z = template.copy();
+ v = template.copy();
+ w = template.copy();
+ p = template.copy();
+ q = template.copy();
+ d = template.copy();
+ s = template.copy();
+ v_tld = template.copy();
+ w_tld = template.copy();
+ y_tld = template.copy();
+ z_tld = template.copy();
+ p_tld = template.copy();
+ }
+
+ public Vector solve(Matrix A, Vector b, Vector x)
+ throws IterativeSolverNotConvergedException {
+ checkSizes(A, b, x);
+
+ double rho = 0, rho_1 = 0, xi = 0, gamma = 1., gamma_1 = 0, theta = 0, theta_1 = 0, eta = -1., delta = 0, ep = 0, beta = 0;
+
+ A.multAdd(-1, x, r.set(b));
+
+ v_tld.set(r);
+ M1.apply(v_tld, y);
+ rho = y.norm(Norm.Two);
+
+ w_tld.set(r);
+ M2.transApply(w_tld, z);
+ xi = z.norm(Norm.Two);
+
+ for (iter.setFirst(); !iter.converged(r, x); iter.next()) {
+
+ if (rho == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "rho", iter);
+
+ if (xi == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "xi", iter);
+
+ v.set(1 / rho, v_tld);
+ y.scale(1 / rho);
+ w.set(1 / xi, w_tld);
+ z.scale(1 / xi);
+
+ delta = z.dot(y);
+
+ if (delta == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "delta", iter);
+
+ M2.apply(y, y_tld);
+ M1.transApply(z, z_tld);
+
+ if (iter.isFirst()) {
+ p.set(y_tld);
+ q.set(z_tld);
+ } else {
+ p.scale(-xi * delta / ep).add(y_tld);
+ q.scale(-rho * delta / ep).add(z_tld);
+ }
+
+ A.mult(p, p_tld);
+
+ ep = q.dot(p_tld);
+
+ if (ep == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "ep", iter);
+
+ beta = ep / delta;
+
+ if (beta == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "beta", iter);
+
+ v_tld.set(-beta, v).add(p_tld);
+ M1.apply(v_tld, y);
+ rho_1 = rho;
+ rho = y.norm(Norm.Two);
+
+ A.transMultAdd(q, w_tld.set(-beta, w));
+ M2.transApply(w_tld, z);
+ xi = z.norm(Norm.Two);
+
+ gamma_1 = gamma;
+ theta_1 = theta;
+ theta = rho / (gamma_1 * beta);
+ gamma = 1 / Math.sqrt(1 + theta * theta);
+
+ if (gamma == 0)
+ throw new IterativeSolverNotConvergedException(
+ NotConvergedException.Reason.Breakdown, "gamma", iter);
+
+ eta = -eta * rho_1 * gamma * gamma / (beta * gamma_1 * gamma_1);
+
+ if (iter.isFirst()) {
+ d.set(eta, p);
+ s.set(eta, p_tld);
+ } else {
+ double val = theta_1 * theta_1 * gamma * gamma;
+ d.scale(val).add(eta, p);
+ s.scale(val).add(eta, p_tld);
+ }
+
+ x.add(d);
+ r.add(-1, s);
+ }
+
+ return x;
+ }
+
+ @Override
+ public void setPreconditioner(Preconditioner M) {
+ super.setPreconditioner(M);
+ M1 = M;
+ M2 = M;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/SSOR.java b/src/main/java/no/uib/cipr/matrix/sparse/SSOR.java
new file mode 100644
index 0000000..4b6a32f
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/SSOR.java
@@ -0,0 +1,201 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * SSOR preconditioner. Uses symmetrical sucessive overrelaxation as a
+ * preconditioner. Meant for symmetrical, positive definite matrices. For best
+ * performance, omega must be carefully chosen (between 0 and 2).
+ */
+public class SSOR implements Preconditioner {
+
+ /**
+ * Overrelaxation parameter for the forward sweep
+ */
+ private double omegaF;
+
+ /**
+ * Overrelaxation parameter for the backwards sweep
+ */
+ private double omegaR;
+
+ /**
+ * Holds a copy of the matrix A in the compressed row format
+ */
+ private final CompRowMatrix F;
+
+ /**
+ * Indices to the diagonal entries of the matrix
+ */
+ private final int[] diagind;
+
+ /**
+ * Temporary vector for holding the half-step state
+ */
+ private final double[] xx;
+
+ /**
+ * True if the reverse (backward) sweep is to be done. Without this, the
+ * method is SOR instead of SSOR
+ */
+ private final boolean reverse;
+
+ /**
+ * Constructor for SSOR
+ *
+ * @param F
+ * Matrix to use internally. It will not be modified, thus the
+ * system matrix may be passed
+ * @param reverse
+ * True to perform a reverse sweep as well as the forward sweep.
+ * If false, this preconditioner becomes the SOR method instead
+ * @param omegaF
+ * Overrelaxation parameter for the forward sweep. Between 0 and
+ * 2.
+ * @param omegaR
+ * Overrelaxation parameter for the backwards sweep. Between 0
+ * and 2.
+ */
+ public SSOR(CompRowMatrix F, boolean reverse, double omegaF, double omegaR) {
+ if (!F.isSquare())
+ throw new IllegalArgumentException(
+ "SSOR only applies to square matrices");
+
+ this.F = F;
+ this.reverse = reverse;
+ setOmega(omegaF, omegaR);
+
+ int n = F.numRows();
+ diagind = new int[n];
+ xx = new double[n];
+ }
+
+ /**
+ * Constructor for SSOR. Uses <code>omega=1</code> with a backwards sweep
+ *
+ * @param F
+ * Matrix to use internally. It will not be modified, thus the
+ * system matrix may be passed
+ */
+ public SSOR(CompRowMatrix F) {
+ this(F, true, 1, 1);
+ }
+
+ /**
+ * Sets the overrelaxation parameters
+ *
+ * @param omegaF
+ * Overrelaxation parameter for the forward sweep. Between 0 and
+ * 2.
+ * @param omegaR
+ * Overrelaxation parameter for the backwards sweep. Between 0
+ * and 2.
+ */
+ public void setOmega(double omegaF, double omegaR) {
+ if (omegaF < 0 || omegaF > 2)
+ throw new IllegalArgumentException("omegaF must be between 0 and 2");
+ if (omegaR < 0 || omegaR > 2)
+ throw new IllegalArgumentException("omegaR must be between 0 and 2");
+
+ this.omegaF = omegaF;
+ this.omegaR = omegaR;
+ }
+
+ public void setMatrix(Matrix A) {
+ F.set(A);
+
+ int n = F.numRows();
+
+ int[] rowptr = F.getRowPointers();
+ int[] colind = F.getColumnIndices();
+
+ // Find the indices to the diagonal entries
+ for (int k = 0; k < n; ++k) {
+ diagind[k] = Arrays.binarySearch(colind, k, rowptr[k],
+ rowptr[k + 1]);
+ if (diagind[k] < 0)
+ throw new RuntimeException("Missing diagonal on row " + (k + 1));
+ }
+ }
+
+ public Vector apply(Vector b, Vector x) {
+ if (!(b instanceof DenseVector) || !(x instanceof DenseVector))
+ throw new IllegalArgumentException("Vectors must be a DenseVectors");
+
+ int[] rowptr = F.getRowPointers();
+ int[] colind = F.getColumnIndices();
+ double[] data = F.getData();
+
+ double[] bd = ((DenseVector) b).getData();
+ double[] xd = ((DenseVector) x).getData();
+
+ int n = F.numRows();
+ System.arraycopy(xd, 0, xx, 0, n);
+
+ // Forward sweep (xd oldest, xx halfiterate)
+ for (int i = 0; i < n; ++i) {
+
+ double sigma = 0;
+ for (int j = rowptr[i]; j < diagind[i]; ++j)
+ sigma += data[j] * xx[colind[j]];
+
+ for (int j = diagind[i] + 1; j < rowptr[i + 1]; ++j)
+ sigma += data[j] * xd[colind[j]];
+
+ sigma = (bd[i] - sigma) / data[diagind[i]];
+
+ xx[i] = xd[i] + omegaF * (sigma - xd[i]);
+ }
+
+ // Stop here if the reverse sweep was not requested
+ if (!reverse) {
+ System.arraycopy(xx, 0, xd, 0, n);
+ return x;
+ }
+
+ // Backward sweep (xx oldest, xd halfiterate)
+ for (int i = n - 1; i >= 0; --i) {
+
+ double sigma = 0;
+ for (int j = rowptr[i]; j < diagind[i]; ++j)
+ sigma += data[j] * xx[colind[j]];
+
+ for (int j = diagind[i] + 1; j < rowptr[i + 1]; ++j)
+ sigma += data[j] * xd[colind[j]];
+
+ sigma = (bd[i] - sigma) / data[diagind[i]];
+
+ xd[i] = xx[i] + omegaR * (sigma - xx[i]);
+ }
+
+ return x;
+ }
+
+ public Vector transApply(Vector b, Vector x) {
+ // Assume a symmetric matrix
+ return apply(b, x);
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/SparseVector.java b/src/main/java/no/uib/cipr/matrix/sparse/SparseVector.java
new file mode 100644
index 0000000..e9717a1
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/SparseVector.java
@@ -0,0 +1,442 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Iterator;
+
+import no.uib.cipr.matrix.AbstractVector;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.VectorEntry;
+
+/**
+ * Sparse vector
+ */
+public class SparseVector extends AbstractVector implements ISparseVector {
+
+ /**
+ * Data
+ */
+ double[] data;
+
+ /**
+ * Indices to data
+ */
+ int[] index;
+
+ /**
+ * How much has been used
+ */
+ int used;
+
+ /**
+ * Constructor for SparseVector.
+ *
+ * @param size
+ * Size of the vector
+ * @param nz
+ * Initial number of non-zeros
+ */
+ public SparseVector(int size, int nz) {
+ super(size);
+ data = new double[nz];
+ index = new int[nz];
+ }
+
+ /**
+ * Constructor for SparseVector, and copies the contents from the supplied
+ * vector.
+ *
+ * @param x
+ * Vector to copy from
+ * @param deep
+ * True if a deep copy is to be made. If the copy is shallow,
+ * <code>x</code> must be a <code>SparseVector</code>
+ */
+ public SparseVector(Vector x, boolean deep) {
+ super(x);
+
+ if (deep) {
+ int nz = Matrices.cardinality(x);
+ data = new double[nz];
+ index = new int[nz];
+ set(x);
+ } else {
+ SparseVector xs = (SparseVector) x;
+ data = xs.getData();
+ index = xs.getIndex();
+ used = xs.getUsed();
+ }
+ }
+
+ /**
+ * Constructor for SparseVector, and copies the contents from the supplied
+ * vector. Zero initial pre-allocation
+ *
+ * @param x
+ * Vector to copy from. A deep copy is made
+ */
+ public SparseVector(Vector x) {
+ this(x, true);
+ }
+
+ /**
+ * Constructor for SparseVector. Zero initial pre-allocation
+ *
+ * @param size
+ * Size of the vector
+ */
+ public SparseVector(int size) {
+ this(size, 0);
+ }
+
+ /**
+ * Constructor for SparseVector
+ *
+ * @param size
+ * Size of the vector
+ * @param index
+ * Indices of the vector
+ * @param data
+ * Entries of the vector
+ * @param deep
+ * True for a deep copy. For shallow copies, the given indices
+ * will be used internally
+ */
+ public SparseVector(int size, int[] index, double[] data, boolean deep) {
+ super(size);
+
+ if (index.length != data.length)
+ throw new IllegalArgumentException("index.length != data.length");
+
+ if (deep) {
+ used = index.length;
+ this.index = index.clone();
+ this.data = data.clone();
+ } else {
+ this.index = index;
+ this.data = data;
+ used = index.length;
+ }
+ }
+
+ /**
+ * Constructor for SparseVector
+ *
+ * @param size
+ * Size of the vector
+ * @param index
+ * The vector indices are copies from this array
+ * @param data
+ * The vector entries are copies from this array
+ */
+ public SparseVector(int size, int[] index, double[] data) {
+ this(size, index, data, true);
+ }
+
+ @Override
+ public void set(int index, double value) {
+ check(index);
+
+ // TODO: should we check against zero when setting zeros?
+
+ int i = getIndex(index);
+ data[i] = value;
+ }
+
+ @Override
+ public void add(int index, double value) {
+ check(index);
+
+ int i = getIndex(index);
+ data[i] += value;
+ }
+
+ @Override
+ public double get(int index) {
+ check(index);
+
+ int in = Arrays.binarySearch(this.index, index, 0, used);
+ if (in >= 0)
+ return data[in];
+ return 0;
+ }
+
+ /**
+ * Tries to find the index. If it is not found, a reallocation is done, and
+ * a new index is returned.
+ */
+ private int getIndex(int ind) {
+
+ // Try to find column index
+ int i = Arrays.binarySearchGreater(index, ind, 0, used);
+
+ // Found
+ if (i < used && index[i] == ind)
+ return i;
+
+ int[] newIndex = index;
+ double[] newData = data;
+
+ // Check available memory
+ if (++used > data.length) {
+
+ // If zero-length, use new length of 1, else double the bandwidth
+ int newLength = data.length != 0 ? data.length << 1 : 1;
+
+ // Copy existing data into new arrays
+ newIndex = new int[newLength];
+ newData = new double[newLength];
+ System.arraycopy(index, 0, newIndex, 0, i);
+ System.arraycopy(data, 0, newData, 0, i);
+ }
+
+ // All ok, make room for insertion
+ System.arraycopy(index, i, newIndex, i + 1, used - i - 1);
+ System.arraycopy(data, i, newData, i + 1, used - i - 1);
+
+ // Put in new structure
+ newIndex[i] = ind;
+ newData[i] = 0.;
+
+ // Update pointers
+ index = newIndex;
+ data = newData;
+
+ // Return insertion index
+ return i;
+ }
+
+ @Override
+ public SparseVector copy() {
+ return new SparseVector(this);
+ }
+
+ @Override
+ public SparseVector zero() {
+ java.util.Arrays.fill(data, 0);
+ used = 0;
+ return this;
+ }
+
+ @Override
+ public SparseVector scale(double alpha) {
+ // Quick return if possible
+ if (alpha == 0)
+ return zero();
+ else if (alpha == 1)
+ return this;
+
+ for (int i = 0; i < used; ++i)
+ data[i] *= alpha;
+
+ return this;
+ }
+
+ @Override
+ public double dot(Vector y) {
+ if (!(y instanceof DenseVector))
+ return super.dot(y);
+
+ checkSize(y);
+
+ double[] yd = ((DenseVector) y).getData();
+
+ double ret = 0;
+ for (int i = 0; i < used; ++i)
+ ret += data[i] * yd[index[i]];
+ return ret;
+ }
+
+ @Override
+ protected double norm1() {
+ double sum = 0;
+ for (int i = 0; i < used; ++i)
+ sum += Math.abs(data[i]);
+ return sum;
+ }
+
+ @Override
+ protected double norm2() {
+ double norm = 0;
+ for (int i = 0; i < used; ++i)
+ norm += data[i] * data[i];
+ return Math.sqrt(norm);
+ }
+
+ @Override
+ protected double norm2_robust() {
+ double scale = 0, ssq = 1;
+ for (int i = 0; i < used; ++i) {
+ if (data[i] != 0) {
+ double absxi = Math.abs(data[i]);
+ if (scale < absxi) {
+ ssq = 1 + ssq * Math.pow(scale / absxi, 2);
+ scale = absxi;
+ } else
+ ssq = ssq + Math.pow(absxi / scale, 2);
+ }
+ }
+ return scale * Math.sqrt(ssq);
+ }
+
+ @Override
+ protected double normInf() {
+ double max = 0;
+ for (int i = 0; i < used; ++i)
+ max = Math.max(Math.abs(data[i]), max);
+ return max;
+ }
+
+ /**
+ * Returns the internal data
+ */
+ public double[] getData() {
+ return data;
+ }
+
+ /**
+ * Returns the indices
+ */
+ public int[] getIndex() {
+ if (used == index.length)
+ return index;
+
+ // could run compact, or return subarray
+ // compact();
+ int [] indices = new int[used];
+ for (int i = 0 ; i < used; i++) {
+ indices[i] = index[i];
+ }
+ return indices;
+ }
+
+ /**
+ * Number of entries used in the sparse structure
+ */
+ public int getUsed() {
+ return used;
+ }
+
+ /**
+ * Compacts the vector
+ */
+ public void compact() {
+ int nz = Matrices.cardinality(this); // catches zero entries
+
+ if (nz < data.length) {
+ int[] newIndex = new int[nz];
+ double[] newData = new double[nz];
+
+ // Copy only non-zero entries
+ for (int i = 0, j = 0; i < data.length; ++i)
+ if (data[i] != 0.) {
+ newIndex[j] = index[i];
+ newData[j] = data[i];
+ j++;
+ }
+
+ data = newData;
+ index = newIndex;
+ used = data.length;
+ }
+ }
+
+ @Override
+ public Iterator<VectorEntry> iterator() {
+ return new SparseVectorIterator();
+ }
+
+ @Override
+ public Vector set(Vector y) {
+ if (!(y instanceof SparseVector))
+ return super.set(y);
+
+ checkSize(y);
+
+ SparseVector yc = (SparseVector) y;
+
+ if (yc.index.length != index.length) {
+ data = new double[yc.data.length];
+ index = new int[yc.data.length];
+ }
+
+ System.arraycopy(yc.data, 0, data, 0, data.length);
+ System.arraycopy(yc.index, 0, index, 0, index.length);
+ used = yc.used;
+
+ return this;
+ }
+
+ /**
+ * Iterator over a sparse vector
+ */
+ private class SparseVectorIterator implements Iterator<VectorEntry> {
+
+ private int cursor;
+
+ private final SparseVectorEntry entry = new SparseVectorEntry();
+
+ public boolean hasNext() {
+ return cursor < used;
+ }
+
+ public VectorEntry next() {
+ entry.update(cursor);
+
+ cursor++;
+
+ return entry;
+ }
+
+ public void remove() {
+ entry.set(0);
+ }
+
+ }
+
+ /**
+ * Entry of a sparse vector
+ */
+ private class SparseVectorEntry implements VectorEntry {
+
+ private int cursor;
+
+ public void update(int cursor) {
+ this.cursor = cursor;
+ }
+
+ public int index() {
+ return index[cursor];
+ }
+
+ public double get() {
+ return data[cursor];
+ }
+
+ public void set(double value) {
+ data[cursor] = value;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/SuperIterator.java b/src/main/java/no/uib/cipr/matrix/sparse/SuperIterator.java
new file mode 100644
index 0000000..41ae65b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/SuperIterator.java
@@ -0,0 +1,158 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Iterator;
+import java.util.List;
+
+/**
+ * An iterator over an array of iterable objects
+ */
+class SuperIterator<T extends Iterable<E>, E> implements
+ Iterator<SuperIterator.SuperIteratorEntry> {
+
+ private List<T> iterable;
+
+ /**
+ * Two iterators. We need the "next" iterator so that hasNext works properly
+ * from one iterable to the next. Using a single iterator won't do
+ */
+ private Iterator<E> current, next;
+
+ private int currentIndex = 0, nextIndex = 0;
+
+ /**
+ * Recyled entry returned from next()
+ */
+ private SuperIteratorEntry<E> entry;
+
+ /**
+ * Constructor for SuperIterator
+ *
+ * @param iterable
+ * Iterable objects to iterate over
+ */
+ public SuperIterator(List<T> iterable) {
+ this.iterable = iterable;
+ entry = new SuperIteratorEntry<E>();
+
+ // Try to be somewhat fault tolerant
+ if (iterable.size() == 0) {
+ current = new DummyIterator();
+ next = new DummyIterator();
+ } else {
+
+ // This moves the next pointer to a non-empty iterable
+ next = iterable.get(nextIndex).iterator();
+ moveNext();
+
+ // Then we move the current pointer in the same way
+ current = iterable.get(currentIndex).iterator();
+ moveCurrent();
+
+ // Finally, move the next one step ahead if possible
+ if (next.hasNext())
+ next.next();
+ }
+ }
+
+ private void moveNext() {
+ while (nextIndex < iterable.size() - 1 && !next.hasNext())
+ next = iterable.get(++nextIndex).iterator();
+ }
+
+ private void moveCurrent() {
+ while (currentIndex < iterable.size() - 1 && !current.hasNext())
+ current = iterable.get(++currentIndex).iterator();
+ }
+
+ public boolean hasNext() {
+ return current.hasNext() || next.hasNext();
+ }
+
+ public SuperIteratorEntry<E> next() {
+ // A wrapped object containing the relevant index and data
+ entry.update(currentIndex, current.next());
+
+ // Move current if necessary
+ moveCurrent();
+
+ // Move the next pointer
+ moveNext();
+ if (next.hasNext())
+ next.next();
+
+ return entry;
+ }
+
+ public void remove() {
+ current.remove();
+ }
+
+ /**
+ * Dummy iterator, for degenerate cases
+ */
+ private class DummyIterator implements Iterator<E> {
+
+ public boolean hasNext() {
+ return false;
+ }
+
+ public E next() {
+ return null;
+ }
+
+ public void remove() {
+ throw new UnsupportedOperationException();
+ }
+ }
+
+ /**
+ * Entry returned from this superiterator
+ */
+ public static class SuperIteratorEntry<F> {
+
+ /**
+ * Index of the iterator which returned this
+ */
+ private int i;
+
+ /**
+ * Object returned
+ */
+ private F o;
+
+ void update(int i, F o) {
+ this.i = i;
+ this.o = o;
+ }
+
+ public int index() {
+ return i;
+ }
+
+ public F get() {
+ return o;
+ }
+
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/UnitLowerCompRowMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/UnitLowerCompRowMatrix.java
new file mode 100644
index 0000000..94eefac
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/UnitLowerCompRowMatrix.java
@@ -0,0 +1,88 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Unit lower triangular CRS matrix. Only used for triangular solves
+ */
+class UnitLowerCompRowMatrix extends AbstractMatrix {
+
+ private int[] rowptr;
+
+ private int[] colind;
+
+ private double[] data;
+
+ private int[] diagind;
+
+ public UnitLowerCompRowMatrix(CompRowMatrix LU, int[] diagind) {
+ super(LU);
+ rowptr = LU.getRowPointers();
+ colind = LU.getColumnIndices();
+ data = LU.getData();
+ this.diagind = diagind;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ if (!(b instanceof DenseVector) || !(x instanceof DenseVector))
+ return super.solve(b, x);
+
+ double[] bd = ((DenseVector) b).getData();
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = 0; i < numRows; ++i) {
+
+ // xi = bi - sum[j<i] Lij * xj
+ double sum = 0;
+ for (int j = rowptr[i]; j < diagind[i]; ++j)
+ sum += data[j] * xd[colind[j]];
+
+ xd[i] = bd[i] - sum;
+ }
+
+ return x;
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ if (!(x instanceof DenseVector))
+ return super.transSolve(b, x);
+
+ x.set(b);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = numRows - 1; i >= 0; --i)
+
+ // At this stage, x[i] is known, so move it over to the right hand
+ // side for the remaining equations
+ for (int j = rowptr[i]; j < diagind[i]; ++j)
+ xd[colind[j]] -= data[j] * xd[i];
+
+ return x;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/UpperCompRowMatrix.java b/src/main/java/no/uib/cipr/matrix/sparse/UpperCompRowMatrix.java
new file mode 100644
index 0000000..c7c22f7
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/UpperCompRowMatrix.java
@@ -0,0 +1,92 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.AbstractMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Vector;
+
+/**
+ * Upper triangular CRS matrix. Only used for triangular solves
+ */
+class UpperCompRowMatrix extends AbstractMatrix {
+
+ private int[] rowptr;
+
+ private int[] colind;
+
+ private double[] data;
+
+ private int[] diagind;
+
+ public UpperCompRowMatrix(CompRowMatrix LU, int[] diagind) {
+ super(LU);
+ rowptr = LU.getRowPointers();
+ colind = LU.getColumnIndices();
+ data = LU.getData();
+ this.diagind = diagind;
+ }
+
+ @Override
+ public Vector solve(Vector b, Vector x) {
+ if (!(b instanceof DenseVector) || !(x instanceof DenseVector))
+ return super.solve(b, x);
+
+ double[] bd = ((DenseVector) b).getData();
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = numRows - 1; i >= 0; --i) {
+
+ // xi = (bi - sum[j>i] Uij * xj) / Uii
+ double sum = 0;
+ for (int j = diagind[i] + 1; j < rowptr[i + 1]; ++j)
+ sum += data[j] * xd[colind[j]];
+
+ xd[i] = (bd[i] - sum) / data[diagind[i]];
+ }
+
+ return x;
+ }
+
+ @Override
+ public Vector transSolve(Vector b, Vector x) {
+ if (!(x instanceof DenseVector))
+ return super.transSolve(b, x);
+
+ x.set(b);
+
+ double[] xd = ((DenseVector) x).getData();
+
+ for (int i = 0; i < numRows; ++i) {
+
+ // Solve for the current entry
+ xd[i] /= data[diagind[i]];
+
+ // Move this known solution over to the right hand side for the
+ // remaining equations
+ for (int j = diagind[i] + 1; j < rowptr[i + 1]; ++j)
+ xd[colind[j]] -= data[j] * xd[i];
+ }
+
+ return x;
+ }
+
+}
diff --git a/src/main/java/no/uib/cipr/matrix/sparse/package.html b/src/main/java/no/uib/cipr/matrix/sparse/package.html
new file mode 100644
index 0000000..7a0475b
--- /dev/null
+++ b/src/main/java/no/uib/cipr/matrix/sparse/package.html
@@ -0,0 +1,63 @@
+<html>
+<body>
+Unstructured sparse matrices and vectors with iterative solvers and
+preconditioners. The classes and interfaces can be grouped as follows:
+<ul>
+ <li><strong>General sparse matrices</strong>
+ <ul>
+ <li><a href="CompRowMatrix.html">CompRowMatrix</a> -
+ Compressed row storage. Generally the best sparse matrix if the non-zero
+ structure is known.</li>
+ <li><a href="CompColMatrix.html">CompColMatrix</a> -
+ Compressed column storage.</li>
+ <li><a href="CompDiagMatrix.html">CompDiagMatrix</a> -
+ Compressed diagonal storage.</li>
+ <li><a href="FlexCompRowMatrix.html">FlexCompRowMatrix</a> -
+ Flexible compressed row storage. Stores each row as a growable sparse
+ vector.</li>
+ <li><a href="FlexCompColMatrix.html">FlexCompColMatrix</a> -
+ Flexible compressed column storage. Stores each column as a growable sparse
+ vector.</li>
+ <li><a href="SparseVector.html">SparseVector</a> -
+ Growable sparse vector.</li>
+ </ul>
+ </li>
+ <li><strong>Iterative solvers</strong>
+ <ul>
+ <li><a href="BiCG.html">BiCG</a> -
+ BiConjugate gradients.</li>
+ <li><a href="BiCGstab.html">BiCGstab</a> -
+ BiConjugate gradients stabilized.</li>
+ <li><a href="CG.html">CG</a> -
+ Conjugate gradients.</li>
+ <li><a href="CGS.html">CGS</a> -
+ Conjugate gradients squared.</li>
+ <li><a href="Chebyshev.html">Chebyshev</a> -
+ The Chebyshev iteration for symmetrical, positive definite matrices.</li>
+ <li><a href="GMRES.html">GMRES</a> -
+ Generalized minimal residual using restart.</li>
+ <li><a href="IR.html">IR</a> -
+ Iterative refinement (Richardson's method).</li>
+ <li><a href="QMR.html">QMR</a> -
+ Quasi-minimal residual.</li>
+ </ul>
+ </li>
+ <li><strong>Preconditioners</strong>
+ <ul>
+ <li><a href="DiagonalPreconditioner.html">DiagonalPreconditioner</a> -
+ Diagonal preconditioning.</li>
+ <li><a href="SSOR.html">SSOR</a> -
+ Symmetrical sucessive overrelaxation.</li>
+ <li><a href="ICC.html">ICC</a> -
+ Incomplete Cholesky without fill-in.</li>
+ <li><a href="ILU.html">ILU</a> -
+ Incomplete LU without fill-in.</li>
+ <li><a href="ILUT.html">ILUT</a> -
+ Incomplete LU with fill-in using a threshold approach.</li>
+ <li><a href="AMG.html">AMG</a> -
+ Algebraic multigrid by smoothed aggregation.</li>
+ </ul>
+ </li>
+</ul>
+</body>
+</html>
diff --git a/src/test/R/no/uib/cipr/matrix/sparse/LinkedSparseMatrixPerf.R b/src/test/R/no/uib/cipr/matrix/sparse/LinkedSparseMatrixPerf.R
new file mode 100644
index 0000000..4f87fe5
--- /dev/null
+++ b/src/test/R/no/uib/cipr/matrix/sparse/LinkedSparseMatrixPerf.R
@@ -0,0 +1,111 @@
+par(cex = 1.5, cex.lab=1.5, cex.axis=1.5, cex.main=1.5, cex.sub=1.5, family="Palatino")
+
+setPdfOut <- function(filename){
+ if (interactive()) {
+ quartz()
+ } else {
+ pdf(filename, width=11, height=8.5)
+ }
+}
+
+logAxis = function(type, lims) {
+ x1 <- floor(log10(lims))
+ pow <- seq(x1[1], x1[2]+1)
+ ticksat <- as.vector(sapply(pow, function(p) (1:10)*10^p))
+ axis(type, 10^pow)
+ axis(type, ticksat, labels=NA, tcl=-0.25, lwd=0, lwd.ticks=1)
+}
+
+
+data = read.csv("LinkedSparseMatrixPerf.csv",
+ col.names=c("n", "m",
+ "denseMem", "denseInit", "denseMult",
+ "sparseMem", "sparseInit", "sparseMult"))
+data$denseInit = data$denseInit / 1000000000
+data$sparseInit = data$sparseInit / 1000000000
+data$denseMult = data$denseMult / 1000000000
+data$sparseMult = data$sparseMult / 1000000000
+
+ms = sort(unique(data$m))
+ns = sort(unique(data$n))
+
+xlim = c(min(ns), max(ns))
+ylim = c(min(data$denseMult), max(data$denseMult))
+setPdfOut("mult.pdf")
+plot(c(), xlim=xlim, xlab="n", ylab="Time (seconds)", log="y", yaxt="n", ylim=ylim, type="n")
+leg = matrix(nrow = 2 * length(ms), ncol = 3)
+for (j in 1:length(ms)) {
+ m = ms[j]
+ print(m)
+ set = data[data$m == m,]
+ avgs = matrix(nrow = length(ns), ncol = 3)
+ for (i in 1:length(ns)) {
+ avgs[i,1] = ns[i]
+ avgs[i,2] = mean(set[set$n == ns[i],]$denseMult)
+ avgs[i,3] = mean(set[set$n == ns[i],]$sparseMult)
+ }
+ col = heat.colors(length(ms))[j]
+ lines(avgs[,1], avgs[,2], lty = 1, col = col)
+ leg[2 * j - 1, 1] = paste("dense", m)
+ leg[2 * j - 1, 2] = col
+ leg[2 * j - 1, 3] = 1
+ lines(avgs[,1], avgs[,3], lty = 2, col = col)
+ leg[2 * j, 1] = paste("sparse", m)
+ leg[2 * j, 2] = col
+ leg[2 * j, 3] = 2
+}
+logAxis(2, ylim)
+
+#legend("bottomright", legend=leg[,1], lty=leg[,3], col=leg[,2], bty="n")
+
+ylim = c(min(data$denseInit), max(data$sparseInit))
+setPdfOut("init.pdf")
+plot(c(), xlim=xlim, xlab="n", ylab="Time (seconds)", log="y", yaxt="n", ylim=ylim, type="n")
+leg = matrix(nrow = 2 * length(ms), ncol = 3)
+for (j in 1:length(ms)) {
+ m = ms[j]
+ print(m)
+ set = data[data$m == m,]
+ avgs = matrix(nrow = length(ns), ncol = 3)
+ for (i in 1:length(ns)) {
+ avgs[i,1] = ns[i]
+ avgs[i,2] = mean(set[set$n == ns[i],]$denseInit)
+ avgs[i,3] = mean(set[set$n == ns[i],]$sparseInit)
+ }
+ col = heat.colors(length(ms))[j]
+ lines(avgs[,1], avgs[,2], lty = 1, col = col)
+ leg[2 * j - 1, 1] = paste("dense", m)
+ leg[2 * j - 1, 2] = col
+ leg[2 * j - 1, 3] = 1
+ lines(avgs[,1], avgs[,3], lty = 2, col = col)
+ leg[2 * j, 1] = paste("sparse", m)
+ leg[2 * j, 2] = col
+ leg[2 * j, 3] = 2
+}
+logAxis(2, ylim)
+
+ylim = c(min(data$sparseMem), max(data$denseMem))
+setPdfOut("mem.pdf")
+plot(c(), xlim=xlim, xlab="n", ylab="Memory (bytes)", log="y", yaxt="n", ylim=ylim, type="n")
+leg = matrix(nrow = 2 * length(ms), ncol = 3)
+for (j in 1:length(ms)) {
+ m = ms[j]
+ print(m)
+ set = data[data$m == m,]
+ avgs = matrix(nrow = length(ns), ncol = 3)
+ for (i in 1:length(ns)) {
+ avgs[i,1] = ns[i]
+ avgs[i,2] = mean(set[set$n == ns[i],]$denseMem)
+ avgs[i,3] = mean(set[set$n == ns[i],]$sparseMem)
+ }
+ col = heat.colors(length(ms))[j]
+ lines(avgs[,1], avgs[,2], lty = 1, col = col)
+ leg[2 * j - 1, 1] = paste("dense", m)
+ leg[2 * j - 1, 2] = col
+ leg[2 * j - 1, 3] = 1
+ lines(avgs[,1], avgs[,3], lty = 2, col = col)
+ leg[2 * j, 1] = paste("sparse", m)
+ leg[2 * j, 2] = col
+ leg[2 * j, 3] = 2
+}
+logAxis(2, ylim)
diff --git a/src/test/java/no/uib/cipr/matrix/BandCholeskyTest.java b/src/test/java/no/uib/cipr/matrix/BandCholeskyTest.java
new file mode 100644
index 0000000..6b5e2e8
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/BandCholeskyTest.java
@@ -0,0 +1,130 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+import no.uib.cipr.matrix.BandCholesky;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.LowerSPDBandMatrix;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.UpperSPDBandMatrix;
+
+/**
+ * Tests the banded Cholesky decomposition
+ */
+public class BandCholeskyTest extends TestCase {
+
+ private LowerSPDBandMatrix L;
+
+ private UpperSPDBandMatrix U;
+
+ private DenseMatrix I;
+
+ private int kl, ku;
+
+ private final int max = 100, bmax = 10;
+
+ public BandCholeskyTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+
+ kl = Math.min(n, Utilities.getInt(bmax));
+ ku = Math.min(n, Utilities.getInt(bmax));
+
+ L = new LowerSPDBandMatrix(n, kl);
+ Utilities.bandPopulate(L, kl, 0);
+ Utilities.addDiagonal(L, 1);
+ while (!Utilities.spd(L))
+ Utilities.addDiagonal(L, 1);
+
+ U = new UpperSPDBandMatrix(n, ku);
+ Utilities.bandPopulate(U, 0, ku);
+ Utilities.addDiagonal(U, 1);
+ while (!Utilities.spd(U))
+ Utilities.addDiagonal(U, 1);
+
+ I = Matrices.identity(n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ L = null;
+ U = null;
+ I = null;
+ }
+
+ public void testLowerBandCholesky() {
+ int n = L.numRows();
+
+ BandCholesky c = new BandCholesky(n, kl, false);
+ c.factor(L.copy());
+
+ c.solve(I);
+
+ Matrix J = I.mult(L, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testUpperBandCholesky() {
+ int n = U.numRows();
+
+ BandCholesky c = new BandCholesky(n, ku, true);
+ c.factor(U.copy());
+
+ c.solve(I);
+
+ Matrix J = I.mult(U, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testLowerBandCholeskyrcond() {
+ int n = L.numRows();
+
+ BandCholesky c = new BandCholesky(n, kl, false);
+ c.factor(L.copy());
+
+ c.rcond(L);
+ }
+
+ public void testUpperBandCholeskyrcond() {
+ int n = U.numRows();
+
+ BandCholesky c = new BandCholesky(n, ku, true);
+ c.factor(U.copy());
+
+ c.rcond(U);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/BandLUTest.java b/src/test/java/no/uib/cipr/matrix/BandLUTest.java
new file mode 100644
index 0000000..a82ea9f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/BandLUTest.java
@@ -0,0 +1,110 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.BandLU;
+import no.uib.cipr.matrix.BandMatrix;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import junit.framework.TestCase;
+
+/**
+ * Tests the banded LU decomposition
+ */
+public class BandLUTest extends TestCase {
+
+ private BandMatrix A;
+
+ private DenseMatrix I;
+
+ private int kl, ku;
+
+ private final int max = 100, bmax = 10;
+
+ public BandLUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+ kl = Math.min(n, Utilities.getInt(bmax));
+ ku = Math.min(n, Utilities.getInt(bmax));
+ A = new BandMatrix(n, kl, kl + ku);
+ Utilities.bandPopulate(A, kl, ku);
+ Utilities.addDiagonal(A, 1);
+ while (Utilities.singular(A))
+ Utilities.addDiagonal(A, 1);
+
+ I = Matrices.identity(n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = null;
+ I = null;
+ }
+
+ public void testBandLU() {
+ int n = A.numRows();
+
+ BandLU lu = new BandLU(n, kl, ku);
+ lu.factor(A.copy());
+
+ lu.solve(I);
+
+ Matrix J = I.mult(A, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testBandLUtranspose() {
+ int n = A.numRows();
+
+ BandLU lu = new BandLU(n, kl, ku);
+ lu.factor(A.copy());
+
+ lu.transSolve(I);
+
+ Matrix J = I.transAmult(A, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testBandLUrcond() {
+ int n = A.numRows();
+
+ BandLU lu = new BandLU(n, kl, ku);
+ lu.factor(A.copy());
+
+ lu.rcond(A, Matrix.Norm.One);
+ lu.rcond(A, Matrix.Norm.Infinity);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/BandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/BandMatrixTest.java
new file mode 100644
index 0000000..0e88a37
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/BandMatrixTest.java
@@ -0,0 +1,60 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.BandMatrix;
+
+/**
+ * Test of BandMatrix
+ */
+public class BandMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public BandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kl = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ int ku = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new BandMatrix(n, kl, ku);
+ Ad = Utilities.bandPopulate(A, kl, ku);
+ }
+
+ @Override
+ public void testTransposeInplace() {
+ BandMatrix B = (BandMatrix) A;
+ if (B.numSubDiagonals() == B.numSuperDiagonals())
+ super.testTransposeInplace();
+ }
+
+ @Override
+ public void testTransMatrixSolve() {
+ // Not supported
+ }
+
+ @Override
+ public void testTransVectorSolve() {
+ // Not supported
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/DenseCholeskyTest.java b/src/test/java/no/uib/cipr/matrix/DenseCholeskyTest.java
new file mode 100644
index 0000000..622fadc
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/DenseCholeskyTest.java
@@ -0,0 +1,126 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseCholesky;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.LowerSPDDenseMatrix;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.UpperSPDDenseMatrix;
+import junit.framework.TestCase;
+
+/**
+ * Tests the dense Cholesky decomposition
+ */
+public class DenseCholeskyTest extends TestCase {
+
+ private LowerSPDDenseMatrix L;
+
+ private UpperSPDDenseMatrix U;
+
+ private DenseMatrix I;
+
+ private final int max = 50;
+
+ public DenseCholeskyTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+
+ L = new LowerSPDDenseMatrix(n);
+ Utilities.lowerPopulate(L);
+ Utilities.addDiagonal(L, 1);
+ while (!Utilities.spd(L))
+ Utilities.addDiagonal(L, 1);
+
+ U = new UpperSPDDenseMatrix(n);
+ Utilities.upperPopulate(U);
+ Utilities.addDiagonal(U, 1);
+ while (!Utilities.spd(U))
+ Utilities.addDiagonal(U, 1);
+
+ I = Matrices.identity(n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ L = null;
+ U = null;
+ I = null;
+ }
+
+ public void testLowerDenseCholesky() {
+ int n = L.numRows();
+
+ DenseCholesky c = new DenseCholesky(n, false);
+ c.factor(L.copy());
+
+ assert I != null;
+ c.solve(I);
+
+ Matrix J = I.mult(L, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testUpperDenseCholesky() {
+ int n = U.numRows();
+
+ DenseCholesky c = new DenseCholesky(n, true);
+ c.factor(U.copy());
+
+ c.solve(I);
+
+ Matrix J = I.mult(U, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testLowerDenseCholeskyrcond() {
+ int n = L.numRows();
+
+ DenseCholesky c = new DenseCholesky(n, false);
+ c.factor(L.copy());
+
+ c.rcond(L);
+ }
+
+ public void testUpperDenseCholeskyrcond() {
+ int n = U.numRows();
+
+ DenseCholesky c = new DenseCholesky(n, true);
+ c.factor(U.copy());
+
+ c.rcond(U);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/DenseLUTest.java b/src/test/java/no/uib/cipr/matrix/DenseLUTest.java
new file mode 100644
index 0000000..8f96539
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/DenseLUTest.java
@@ -0,0 +1,120 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+
+/**
+ * Tests the dense LU decomposition
+ */
+public class DenseLUTest extends TestCase {
+
+ /**
+ * Matrix to decompose
+ */
+ private DenseMatrix A;
+
+ private DenseMatrix I;
+
+ private final int max = 100;
+
+ public DenseLUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new DenseMatrix(n, n);
+ Utilities.populate(A);
+ Utilities.addDiagonal(A, 1);
+ while (Utilities.singular(A))
+ Utilities.addDiagonal(A, 1);
+
+ I = Matrices.identity(n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = null;
+ I = null;
+ }
+
+ public void testDenseLU() {
+ int n = A.numRows();
+ DenseLU lu = new DenseLU(n, n);
+ lu.factor(A.copy());
+
+ lu.solve(I);
+
+ Matrix J = I.mult(A, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testDenseLUtranspose() {
+ int n = A.numRows();
+ DenseLU lu = new DenseLU(n, n);
+ lu.factor(A.copy());
+
+ lu.transSolve(I);
+
+ Matrix J = I.transAmult(A, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testDenseLUrcond() {
+ int n = A.numRows();
+ DenseLU lu = new DenseLU(n, n);
+ lu.factor(A.copy());
+
+ lu.rcond(A, Matrix.Norm.One);
+ lu.rcond(A, Matrix.Norm.Infinity);
+ }
+
+ public void testDensePLU() {
+ Matrix m = new DenseMatrix(new double[][]{
+ {2, -1, -2},
+ {-4, 6, 3},
+ {-4, -2, 8}
+ });
+ DenseLU dlu = DenseLU.factorize(m);
+
+ Matrix p = dlu.getP();
+ Matrix l = dlu.getL();
+ Matrix u = dlu.getU();
+
+ Matrix lu = l.mult(u, new DenseMatrix(3, 3));
+ Matrix x = p.mult(lu, new DenseMatrix(3, 3));
+
+ MatrixTestAbstract.assertEquals(m, x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/DenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/DenseMatrixTest.java
new file mode 100644
index 0000000..4341ed8
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/DenseMatrixTest.java
@@ -0,0 +1,123 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+
+/**
+ * Test of a dense matrix
+ */
+public class DenseMatrixTest extends MatrixTestAbstract {
+
+ public DenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ A = new DenseMatrix(n, m);
+ Ad = Utilities.populate(A);
+ }
+
+ @Override
+ public void testMatrixSolve() {
+ if (A.isSquare())
+ super.testMatrixSolve();
+ }
+
+ @Override
+ public void testTransMatrixSolve() {
+ if (A.isSquare())
+ super.testTransMatrixSolve();
+ }
+
+ @Override
+ public void testTransVectorSolve() {
+ if (A.isSquare())
+ super.testTransVectorSolve();
+ }
+
+ @Override
+ public void testVectorSolve() {
+ if (A.isSquare())
+ super.testVectorSolve();
+ }
+
+ public void testIssue13(){
+ Vector bv = Matrices.random(100);
+ Matrix am = Matrices.random(100, 50);
+ Vector xv = new DenseVector(am.numColumns());
+ for (int x = 0; x < am.numColumns(); x++) {
+ xv.set(x, 1);
+ }
+ xv = Matrices.random(xv.size());
+ xv = am.solve(bv, xv);
+ }
+
+ public void testIssue32(){
+
+ // The issue here is that we should not allow matrices with more than
+ // Integer.MAX_VALUE entries.
+ boolean exceptionThrown = false;
+ try
+ {
+ Matrix m = new DenseMatrix(Integer.MAX_VALUE, 2);
+ }
+ catch (IllegalArgumentException e)
+ {
+ exceptionThrown = true;
+ }
+ finally
+ {
+ assertTrue(exceptionThrown);
+ }
+
+ exceptionThrown = false;
+ try
+ {
+ Matrix m = new DenseMatrix(Integer.MAX_VALUE, 3);
+ }
+ catch (IllegalArgumentException e)
+ {
+ exceptionThrown = true;
+ }
+ finally
+ {
+ assertTrue(exceptionThrown);
+ }
+
+ exceptionThrown = false;
+ try
+ {
+ Matrix m = new DenseMatrix(Integer.MAX_VALUE - 10, 3);
+ }
+ catch (IllegalArgumentException e)
+ {
+ exceptionThrown = true;
+ }
+ finally
+ {
+ assertTrue(exceptionThrown);
+ }
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/DenseVectorSubTest.java b/src/test/java/no/uib/cipr/matrix/DenseVectorSubTest.java
new file mode 100644
index 0000000..41f61b1
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/DenseVectorSubTest.java
@@ -0,0 +1,23 @@
+package no.uib.cipr.matrix;
+
+import java.util.Random;
+
+/**
+ * @author Sam Halliday
+ */
+public class DenseVectorSubTest extends VectorTestAbstract {
+
+ public DenseVectorSubTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ DenseVector wrapped = new DenseVector(n * 10);
+ int offset = new Random().nextInt(n * 9);
+ x = new DenseVectorSub(wrapped, offset, n);
+ xd = Utilities.populate(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/DenseVectorTest.java b/src/test/java/no/uib/cipr/matrix/DenseVectorTest.java
new file mode 100644
index 0000000..0b41405
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/DenseVectorTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseVector;
+
+/**
+ * Test of DenseVector
+ */
+public class DenseVectorTest extends VectorTestAbstract {
+
+ public DenseVectorTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ x = new DenseVector(n);
+ xd = Utilities.populate(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LQTest.java b/src/test/java/no/uib/cipr/matrix/LQTest.java
new file mode 100644
index 0000000..778ff28
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LQTest.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.LQ;
+import no.uib.cipr.matrix.Matrix;
+
+/**
+ * LQ test
+ */
+public class LQTest extends OrthogonalTestAbstract {
+
+ public LQTest(String arg0) {
+ super(arg0);
+ }
+
+ public void testStaticFactorize() {
+ assertEquals(A, LQ.factorize(A));
+ }
+
+ public void testRepeatStaticFactorize() {
+ assertEquals(A, LQ.factorize(A));
+ assertEquals(A, LQ.factorize(A));
+ }
+
+ public void testFactor() {
+ LQ lq = new LQ(A.numRows(), A.numColumns());
+ assertEquals(A, lq.factor(new DenseMatrix(A)));
+ }
+
+ public void testRepeatFactor() {
+ LQ lq = new LQ(A.numRows(), A.numColumns());
+ lq.factor(new DenseMatrix(A));
+ assertEquals(A, lq);
+ lq.factor(new DenseMatrix(A));
+ assertEquals(A, lq);
+ }
+
+ public void testStaticFactorizeNonSquare() {
+ assertEquals(Ac, LQ.factorize(Ac));
+ }
+
+ public void testRepeatStaticFactorizeNonSquare() {
+ assertEquals(Ac, LQ.factorize(Ac));
+ assertEquals(Ac, LQ.factorize(Ac));
+ }
+
+ public void testFactorNonSquare() {
+ LQ lq = new LQ(Ac.numRows(), Ac.numColumns());
+ assertEquals(Ac, lq.factor(new DenseMatrix(Ac)));
+ }
+
+ public void testRepeatFactorNonSquare() {
+ LQ lq = new LQ(Ac.numRows(), Ac.numColumns());
+ lq.factor(new DenseMatrix(Ac));
+ assertEquals(Ac, lq);
+ lq.factor(new DenseMatrix(Ac));
+ assertEquals(Ac, lq);
+ }
+
+ private void assertEquals(Matrix A, LQ lq) {
+ assertEquals(A, lq.getL().mult(lq.getQ(), A.copy().zero()));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSPDBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSPDBandMatrixTest.java
new file mode 100644
index 0000000..cbdd77d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSPDBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSPDBandMatrix;
+
+/**
+ * Test of LowerSPDBandMatrix
+ */
+public class LowerSPDBandMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public LowerSPDBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new LowerSPDBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, kd, kd);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSPDDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSPDDenseMatrixTest.java
new file mode 100644
index 0000000..2bdeac9
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSPDDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSPDDenseMatrix;
+
+/**
+ * Test of LowerSPDDenseMatrix
+ */
+public class LowerSPDDenseMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public LowerSPDDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerSPDDenseMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSPDPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSPDPackMatrixTest.java
new file mode 100644
index 0000000..0bdfffc
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSPDPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSPDPackMatrix;
+
+/**
+ * Test of LowerSPDPackMatrix
+ */
+public class LowerSPDPackMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public LowerSPDPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerSPDPackMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSymmBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSymmBandMatrixTest.java
new file mode 100644
index 0000000..4934471
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSymmBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmBandMatrix;
+
+/**
+ * Test of LowerSymmBandMatrix
+ */
+public class LowerSymmBandMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public LowerSymmBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new LowerSymmBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, kd, kd);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSymmDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSymmDenseMatrixTest.java
new file mode 100644
index 0000000..7f50432
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSymmDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmDenseMatrix;
+
+/**
+ * Test of LowerSymmDenseMatrix
+ */
+public class LowerSymmDenseMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public LowerSymmDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerSymmDenseMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerSymmPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerSymmPackMatrixTest.java
new file mode 100644
index 0000000..819580b
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerSymmPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmPackMatrix;
+
+/**
+ * Test of LowerSymmPackMatrix
+ */
+public class LowerSymmPackMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public LowerSymmPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerSymmPackMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerTriangBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerTriangBandMatrixTest.java
new file mode 100644
index 0000000..e8cff11
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerTriangBandMatrixTest.java
@@ -0,0 +1,45 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerTriangBandMatrix;
+
+/**
+ * Test of LowerTriangBandMatrix
+ */
+public class LowerTriangBandMatrixTest extends TriangMatrixTestAbstract {
+
+ public LowerTriangBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new LowerTriangBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, kd, 0);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerTriangDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerTriangDenseMatrixTest.java
new file mode 100644
index 0000000..b5f2b83
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerTriangDenseMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerTriangDenseMatrix;
+
+/**
+ * Test of LowerTriangDenseMatrix
+ */
+public class LowerTriangDenseMatrixTest extends TriangMatrixTestAbstract {
+
+ public LowerTriangDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerTriangDenseMatrix(n);
+ Ad = Utilities.lowerPopulate(A);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/LowerTriangPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/LowerTriangPackMatrixTest.java
new file mode 100644
index 0000000..efed6b0
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/LowerTriangPackMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerTriangPackMatrix;
+
+/**
+ * Test of LowerTriangPackMatrix
+ */
+public class LowerTriangPackMatrixTest extends TriangMatrixTestAbstract {
+
+ public LowerTriangPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new LowerTriangPackMatrix(n);
+ Ad = Utilities.lowerPopulate(A);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/MatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/MatrixTestAbstract.java
new file mode 100644
index 0000000..2094950
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/MatrixTestAbstract.java
@@ -0,0 +1,1224 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+
+/**
+ * Tests a matrix
+ */
+public abstract class MatrixTestAbstract extends TestCase {
+
+ /**
+ * Matrix to test
+ */
+ protected Matrix A;
+
+ /**
+ * Jagged array version of A
+ */
+ protected double[][] Ad;
+
+ /**
+ * Matrix of the same size as A, dense and non-dense
+ */
+ protected Matrix Bdense, B;
+
+ /**
+ * Contents of B
+ */
+ protected double[][] Bd;
+
+ /**
+ * Non-dense vectors with size equal the number of rows in A
+ */
+ protected Vector xR, yR;
+
+ /**
+ * Non-dense vectors with size equal the number of columns in A
+ */
+ protected Vector xC, yC;
+
+ /**
+ * Dense vectors with size equal the number of rows in A
+ */
+ protected Vector xDenseR, yDenseR;
+
+ /**
+ * Dense vectors with size equal the number of columns in A
+ */
+ protected Vector xDenseC, yDenseC;
+
+ /**
+ * Contents of the vectors
+ */
+ protected double[] xdR, ydR, xdC, ydC;
+
+ /**
+ * Tolerance for floating-point comparisons
+ */
+ protected double tol = 1e-4;
+
+ /**
+ * Maximum matrix size, to avoid too slow tests
+ */
+ protected int max = 100;
+
+ /**
+ * Constructor for MatrixTestAbstract
+ */
+ public MatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ createPrimary();
+ createAuxillerary();
+ }
+
+ protected abstract void createPrimary() throws Exception;
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = B = Bdense = null;
+ Ad = Bd = null;
+ xC = xDenseC = xDenseR = xR = yC = yDenseC = yDenseR = yR = null;
+ xdC = xdR = ydC = ydR = null;
+ }
+
+ /**
+ * Called after setUp() to create additional datastructures
+ */
+ protected void createAuxillerary() {
+ Bdense = Matrices.random(A.numRows(), A.numColumns());
+ B = Matrices.synchronizedMatrix(Bdense.copy());
+ Bd = Matrices.getArray(B);
+
+ xDenseC = Matrices.random(A.numColumns());
+ yDenseC = Matrices.random(A.numColumns());
+
+ xDenseR = Matrices.random(A.numRows());
+ yDenseR = Matrices.random(A.numRows());
+
+ xC = Matrices.synchronizedVector(xDenseC);
+ yC = Matrices.synchronizedVector(yDenseC);
+
+ xR = Matrices.synchronizedVector(xDenseR);
+ yR = Matrices.synchronizedVector(yDenseR);
+
+ xdC = Matrices.getArray(xC);
+ ydC = Matrices.getArray(yC);
+
+ xdR = Matrices.getArray(xR);
+ ydR = Matrices.getArray(yR);
+ }
+
+ public void testMatrixRank2Dense() {
+ if (A.isSquare()) {
+ int n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(A.numRows(), n), C = Matrices.random(A
+ .numRows(), n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.rank2(alpha, B, C);
+ rank2(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixRank2() {
+ if (A.isSquare()) {
+ int n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(A.numRows(),
+ n)), C = Matrices.synchronizedMatrix(Matrices.random(A
+ .numRows(), n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.rank2(alpha, B, C);
+ rank2(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixTransRank2Dense() {
+ if (A.isSquare()) {
+ int n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(n, A.numColumns()), C = Matrices.random(
+ n, A.numColumns());
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.transRank2(alpha, B, C);
+ transRank2(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixTransRank2() {
+ if (A.isSquare()) {
+ int n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(n, A
+ .numColumns())), C = Matrices.synchronizedMatrix(Matrices
+ .random(n, A.numColumns()));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.transRank2(alpha, B, C);
+ transRank2(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixRank1Dense() {
+ if (A.isSquare()) {
+ Matrix C = Matrices.random(A.numRows(), A.numColumns());
+ double[][] Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.rank1(alpha, C);
+ rank1(Ad, alpha, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixRank1() {
+ if (A.isSquare()) {
+ Matrix C = Matrices.synchronizedMatrix(Matrices.random(A.numRows(),
+ A.numColumns()));
+ double[][] Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.rank1(alpha, C);
+ rank1(Ad, alpha, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixTransRank1Dense() {
+ if (A.isSquare()) {
+ Matrix C = Matrices.random(A.numRows(), A.numColumns());
+ double[][] Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.transRank1(alpha, C);
+ transRank1(Ad, alpha, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixTransRank1() {
+ if (A.isSquare()) {
+ Matrix C = Matrices.synchronizedMatrix(Matrices.random(A.numRows(),
+ A.numColumns()));
+ double[][] Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ A = A.transRank1(alpha, C);
+ transRank1(Ad, alpha, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Cd, C);
+ }
+ }
+
+ public void testMatrixMultDense() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(k, n), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.mult(alpha, B, C);
+ Cd = mult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixMult() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(k, n)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.mult(alpha, B, C);
+ Cd = mult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransAmultDense() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(k, n), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transAmult(alpha, B, C);
+ Cd = transAmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransAmult() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(k, n)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transAmult(alpha, B, C);
+ Cd = transAmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransABmultDense() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(n, k), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transABmult(alpha, B, C);
+ Cd = transABmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransABmult() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(n, k)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transABmult(alpha, B, C);
+ Cd = transABmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransBmultDense() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(n, k), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transBmult(alpha, B, C);
+ Cd = transBmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransBmult() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(n, k)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transBmult(alpha, B, C);
+ Cd = transBmult(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixMultAddDense() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(k, n), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.multAdd(alpha, B, C);
+ Cd = multAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixMultAdd() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(k, n)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.multAdd(alpha, B, C);
+ Cd = multAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransAmultAddDense() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(k, n), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transAmultAdd(alpha, B, C);
+ Cd = transAmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransAmultAdd() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(k, n)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transAmultAdd(alpha, B, C);
+ Cd = transAmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransABmultAddDense() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(n, k), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transABmultAdd(alpha, B, C);
+ Cd = transABmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransABmultAdd() {
+ int m = A.numColumns(), k = A.numRows(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(n, k)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transABmultAdd(alpha, B, C);
+ Cd = transABmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransBmultAddDense() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.random(n, k), C = Matrices.random(m, n);
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transBmultAdd(alpha, B, C);
+ Cd = transBmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ public void testMatrixTransBmultAdd() {
+ int m = A.numRows(), k = A.numColumns(), n = Utilities.getInt(1, max);
+ Matrix B = Matrices.synchronizedMatrix(Matrices.random(n, k)), C = Matrices
+ .synchronizedMatrix(Matrices.random(m, n));
+ double[][] Bd = Matrices.getArray(B), Cd = Matrices.getArray(C);
+ double alpha = Math.random();
+
+ C = A.transBmultAdd(alpha, B, C);
+ Cd = transBmultAdd(Ad, alpha, Bd, Cd);
+
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ assertEquals(Cd, C);
+ }
+
+ protected double[][] rank2(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ return transBmultAdd(Bd, alpha, Cd, transBmultAdd(Cd, alpha, Bd, Ad));
+ }
+
+ protected double[][] transRank2(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ return transAmultAdd(Bd, alpha, Cd, transAmultAdd(Cd, alpha, Bd, Ad));
+ }
+
+ protected double[][] rank1(double[][] Ad, double alpha, double[][] Cd) {
+ return transBmultAdd(Cd, alpha, Cd, Ad);
+ }
+
+ protected double[][] transRank1(double[][] Ad, double alpha, double[][] Cd) {
+ return transAmultAdd(Cd, alpha, Cd, Ad);
+ }
+
+ public void testVectorRank2Dense() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank2(alpha, xdR, ydR), A.rank2(alpha, xDenseR,
+ yDenseR));
+ }
+ }
+
+ public void testVectorRank2() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank2(alpha, xdR, ydR), A.rank2(alpha, xR, yR));
+ }
+ }
+
+ public void testVectorRank1Dense() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank1(alpha, xdR, ydR), A.rank1(alpha, xDenseR,
+ yDenseR));
+ }
+ }
+
+ public void testVectorRank1() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank1(alpha, xdR, ydR), A.rank1(alpha, xR, yR));
+ }
+ }
+
+ protected double[][] rank2(double alpha, double[] xd, double[] yd) {
+ rank1(alpha, xd, yd);
+ rank1(alpha, yd, xd);
+ return Ad;
+ }
+
+ protected double[][] rank1(double alpha, double[] xd, double[] yd) {
+ for (int i = 0; i < xd.length; ++i)
+ for (int j = 0; j < yd.length; ++j)
+ Ad[i][j] += alpha * xd[i] * yd[j];
+ return Ad;
+ }
+
+ public void testVectorTransMultAddDense() {
+ double alpha = Math.random();
+ assertEquals(transMultAdd(alpha, xdR, ydC), A.transMultAdd(alpha,
+ xDenseR, yDenseC));
+ assertEquals(Ad, A);
+ assertEquals(xdR, xDenseR);
+ assertEquals(ydC, yDenseC);
+ }
+
+ public void testVectorTransMultAdd() {
+ double alpha = Math.random();
+ assertEquals(transMultAdd(alpha, xdR, ydC), A.transMultAdd(alpha, xR,
+ yC));
+ assertEquals(Ad, A);
+ assertEquals(xdR, xR);
+ assertEquals(ydC, yC);
+ }
+
+ protected double[] transMultAdd(double alpha, double[] xd, double[] yd) {
+ int rows = Ad.length, cols = 0;
+ if (rows > 0)
+ cols = Ad[0].length;
+
+ for (int j = 0; j < cols; ++j) {
+ double dot = 0;
+ for (int i = 0; i < rows; ++i)
+ dot += Ad[i][j] * xd[i];
+ yd[j] += alpha * dot;
+ }
+
+ return yd;
+ }
+
+ public void testVectorMultDense() {
+ double alpha = Math.random();
+ assertEquals(mult(alpha, xdC, ydR), A.mult(alpha, xDenseC, yDenseR));
+ assertEquals(Ad, A);
+ assertEquals(xdC, xDenseC);
+ assertEquals(ydR, yDenseR);
+ }
+
+ public void testVectorMult() {
+ double alpha = Math.random();
+ assertEquals(mult(alpha, xdC, ydR), A.mult(alpha, xC, yR));
+ assertEquals(Ad, A);
+ assertEquals(xdC, xC);
+ assertEquals(ydR, yR);
+ }
+
+ protected double[] mult(double alpha, double[] xd, double[] yd) {
+ for (int i = 0; i < Ad.length; ++i) {
+ double dot = 0;
+ for (int j = 0; j < Ad[i].length; ++j)
+ dot += Ad[i][j] * xd[j];
+ yd[i] = alpha * dot;
+ }
+ return yd;
+ }
+
+ public void testVectorMultAddDense() {
+ double alpha = Math.random();
+ assertEquals(multAdd(Ad, alpha, xdC, ydR), A.multAdd(alpha, xDenseC,
+ yDenseR));
+ assertEquals(Ad, A);
+ assertEquals(xdC, xDenseC);
+ assertEquals(ydR, yDenseR);
+ }
+
+ public void testVectorMultAdd() {
+ double alpha = Math.random();
+ assertEquals(multAdd(Ad, alpha, xdC, ydR), A.multAdd(alpha, xC, yR));
+ assertEquals(Ad, A);
+ assertEquals(xdC, xC);
+ assertEquals(ydR, yR);
+ }
+
+ protected double[] multAdd(double[][] Ad, double alpha, double[] xd,
+ double[] yd) {
+ for (int i = 0; i < Ad.length; ++i) {
+ double dot = 0;
+ for (int j = 0; j < Ad[i].length; ++j)
+ dot += Ad[i][j] * xd[j];
+ yd[i] += alpha * dot;
+ }
+
+ return yd;
+ }
+
+ protected double[][] mult(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ int m = Cd.length, n = 0, k = Bd.length;
+ if (k > 0)
+ n = Bd[0].length;
+
+ Utilities.zero(Cd);
+
+ for (int j = 0; j < n; ++j)
+ for (int l = 0; l < k; ++l)
+ for (int i = 0; i < m; ++i)
+ Cd[i][j] += alpha * Ad[i][l] * Bd[l][j];
+
+ return Cd;
+ }
+
+ protected double[][] transAmult(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ int m = Cd.length, n = 0, k = Bd.length;
+ if (k > 0)
+ n = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int i = 0; i < m; ++i) {
+ double temp = 0;
+ for (int l = 0; l < k; ++l)
+ temp += Ad[l][i] * Bd[l][j];
+ Cd[i][j] = alpha * temp;
+ }
+
+ return Cd;
+ }
+
+ protected double[][] transBmult(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ int m = Cd.length, n = Bd.length, k = 0;
+ if (n > 0)
+ k = Bd[0].length;
+
+ Utilities.zero(Cd);
+
+ for (int j = 0; j < n; ++j) {
+ for (int l = 0; l < k; ++l)
+ for (int i = 0; i < m; ++i)
+ Cd[i][j] += alpha * Ad[i][l] * Bd[j][l];
+ }
+
+ return Cd;
+ }
+
+ protected double[][] transABmult(double[][] Ad, double alpha,
+ double[][] Bd, double[][] Cd) {
+ int m = Cd.length, n = Bd.length, k = 0;
+ if (n > 0)
+ k = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int i = 0; i < m; ++i) {
+ double temp = 0;
+ for (int l = 0; l < k; ++l)
+ temp += Ad[l][i] * Bd[j][l];
+ Cd[i][j] = alpha * temp;
+ }
+
+ return Cd;
+ }
+
+ protected double[][] multAdd(double[][] Ad, double alpha, double[][] Bd,
+ double[][] Cd) {
+ int m = Cd.length, n = 0, k = Bd.length;
+ if (k > 0)
+ n = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int l = 0; l < k; ++l)
+ for (int i = 0; i < m; ++i)
+ Cd[i][j] += alpha * Ad[i][l] * Bd[l][j];
+
+ return Cd;
+ }
+
+ protected double[][] transAmultAdd(double[][] Ad, double alpha,
+ double[][] Bd, double[][] Cd) {
+ int m = Cd.length, n = 0, k = Bd.length;
+ if (k > 0)
+ n = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int i = 0; i < m; ++i) {
+ double temp = 0;
+ for (int l = 0; l < k; ++l)
+ temp += Ad[l][i] * Bd[l][j];
+ Cd[i][j] += alpha * temp;
+ }
+
+ return Cd;
+ }
+
+ protected double[][] transBmultAdd(double[][] Ad, double alpha,
+ double[][] Bd, double[][] Cd) {
+ int m = Cd.length, n = Bd.length, k = 0;
+ if (n > 0)
+ k = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int l = 0; l < k; ++l)
+ for (int i = 0; i < m; ++i)
+ Cd[i][j] += alpha * Ad[i][l] * Bd[j][l];
+
+ return Cd;
+ }
+
+ protected double[][] transABmultAdd(double[][] Ad, double alpha,
+ double[][] Bd, double[][] Cd) {
+ int m = Cd.length, n = Bd.length, k = 0;
+ if (n > 0)
+ k = Bd[0].length;
+
+ for (int j = 0; j < n; ++j)
+ for (int i = 0; i < m; ++i) {
+ double temp = 0;
+ for (int l = 0; l < k; ++l)
+ temp += Ad[l][i] * Bd[j][l];
+ Cd[i][j] += alpha * temp;
+ }
+
+ return Cd;
+ }
+
+ /**
+ * Tests <code>A = A + alpha*B</code>
+ */
+ public void testRandomMatrixAdd() {
+ double alpha = Math.random();
+ A = A.add(alpha, B);
+ add(Ad, alpha, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = A + B</code>
+ */
+ public void testMatrixAdd() {
+ A = A.add(B);
+ add(Ad, 1, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = A + 1*B</code>
+ */
+ public void testOneMatrixAdd() {
+ A = A.add(1, B);
+ add(Ad, 1, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = A + 0*B</code>
+ */
+ public void testZeroMatrixAdd() {
+ A = A.add(0, B);
+ add(Ad, 0, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = alpha*B</code>
+ */
+ public void testRandomMatrixSet() {
+ double alpha = Math.random();
+ A = A.set(alpha, B);
+ set(Ad, alpha, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = B</code>
+ */
+ public void testMatrixSet() {
+ A = A.set(B);
+ set(Ad, 1, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = 1*B</code>
+ */
+ public void testOneMatrixSet() {
+ A = A.set(1, B);
+ set(Ad, 1, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Tests <code>A = 0*B</code>
+ */
+ public void testZeroMatrixSet() {
+ A = A.set(0, B);
+ set(Ad, 0, Bd);
+ assertEquals(Ad, A);
+ assertEquals(Bd, B);
+ }
+
+ /**
+ * Checks transpose
+ */
+ public void testTranspose() {
+ Matrix At = Matrices.random(A.numColumns(), A.numRows());
+ assertEquals(transpose(), A.transpose(At));
+ }
+
+ protected void set(double[][] A, double alpha, double[][] B) {
+ for (int i = 0; i < A.length; ++i)
+ for (int j = 0; j < A[i].length; ++j)
+ A[i][j] = alpha * B[i][j];
+ }
+
+ protected void add(double[][] A, double alpha, double[][] B) {
+ for (int i = 0; i < A.length; ++i)
+ for (int j = 0; j < A[i].length; ++j)
+ A[i][j] += alpha * B[i][j];
+ }
+
+ protected double[][] transpose() {
+ if (Ad.length == 0)
+ return new double[0][0];
+
+ double[][] Adt = new double[Ad[0].length][Ad.length];
+ for (int i = 0; i < Ad.length; ++i)
+ for (int j = 0; j < Ad[i].length; ++j)
+ Adt[j][i] = Ad[i][j];
+ return Adt;
+ }
+
+ /**
+ * Test of direct matrix solver
+ */
+ public void testMatrixSolve() {
+ while (true) {
+ try {
+ Matrix B = Matrices.random(A.numRows(), A.numColumns());
+ Matrix X = Matrices.random(A.numRows(), A.numColumns());
+ X = A.solve(B, X);
+
+ Matrix Y = A.multAdd(X, X.copy().set(-1, B));
+ assertEquals(0, Y.norm(Matrix.Norm.Frobenius), tol);
+ assertEquals(Ad, A);
+ return;
+ } catch (MatrixSingularException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ } catch (MatrixNotSPDException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+ }
+ }
+
+ /**
+ * Test of direct transpose matrix solver
+ */
+ public void testTransMatrixSolve() {
+ while (true) {
+ try {
+ Matrix B = Matrices.random(A.numRows(), A.numColumns());
+ Matrix X = Matrices.random(A.numRows(), A.numColumns());
+ X = A.transSolve(B, X);
+
+ Matrix Y = A.transAmultAdd(X, X.copy().set(-1, B));
+ assertEquals(0, Y.norm(Matrix.Norm.Frobenius), tol);
+ assertEquals(Ad, A);
+ return;
+ } catch (MatrixSingularException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ } catch (MatrixNotSPDException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+ }
+ }
+
+ /**
+ * Test of direct vector solver
+ */
+ public void testVectorSolve() {
+ while (true) {
+ try {
+ Vector b = Matrices.random(A.numRows());
+ Vector x = Matrices.random(A.numRows());
+ x = A.solve(b, x);
+
+ Vector y = A.multAdd(-1, x, x.copy().set(b));
+ assertEquals(0, y.norm(Vector.Norm.Two), tol);
+ assertEquals(Ad, A);
+ return;
+ } catch (MatrixSingularException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ } catch (MatrixNotSPDException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+ }
+ }
+
+ /**
+ * Test of direct transpose vector solver
+ */
+ public void testTransVectorSolve() {
+ while (true) {
+ try {
+ Vector b = Matrices.random(A.numRows());
+ Vector x = Matrices.random(A.numRows());
+ x = A.transSolve(b, x);
+
+ Vector y = A.transMultAdd(-1, x, x.copy().set(b));
+ assertEquals(0, y.norm(Vector.Norm.Two), tol);
+ assertEquals(Ad, A);
+ return;
+ } catch (MatrixSingularException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ } catch (MatrixNotSPDException e) {
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+ }
+ }
+
+ /**
+ * Test additions using iterators
+ */
+ public void testAdd() {
+ double alpha = Math.random();
+ for (MatrixEntry e : A) {
+ A.add(e.row(), e.column(), alpha);
+ A.add(e.row(), e.column(), -alpha);
+ }
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Checks that copy is deep, not reference
+ */
+ public void testCopy() {
+ Matrix Ac = A.copy();
+ A = A.zero();
+ assertEquals(Ad, Ac);
+ }
+
+ /**
+ * Test iterator get
+ */
+ public void testIterator() {
+ double[][] Ac = new double[A.numRows()][A.numColumns()];
+ for (MatrixEntry e : A)
+ Ac[e.row()][e.column()] = e.get();
+ assertEquals(Ad, Ac);
+ }
+
+ /**
+ * Test iterator set
+ */
+ public void testIteratorSet() {
+ double alpha = Math.random();
+ for (MatrixEntry e : A)
+ e.set(e.get() * alpha);
+ assertEquals(scale(alpha), A);
+ }
+
+ /**
+ * Test iterator read and write
+ */
+ public void testIteratorSetGet() {
+ double alpha = Math.random();
+ double[][] Ac = new double[A.numRows()][A.numColumns()];
+ for (MatrixEntry e : A) {
+ Ac[e.row()][e.column()] = e.get();
+ e.set(alpha * e.get());
+ e.set(e.get() / alpha);
+ }
+ assertEquals(Ad, Ac);
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Checks zero()
+ */
+ public void testZero() {
+ assertEquals(zero(), A.zero());
+ }
+
+ protected double[][] zero() {
+ for (int i = 0; i < Ad.length; ++i)
+ for (int j = 0; j < Ad[i].length; ++j)
+ Ad[i][j] = 0;
+ return Ad;
+ }
+
+ /**
+ * Cardinality computation
+ */
+ public void testCardinality() {
+ assertEquals(Matrices.cardinality(A), cardinality());
+ }
+
+ protected int cardinality() {
+ int nz = 0;
+ for (int i = 0; i < Ad.length; ++i)
+ for (int j = 0; j < Ad[i].length; ++j)
+ if (Ad[i][j] != 0.)
+ nz++;
+ return nz;
+ }
+
+ /**
+ * Checks in-place transpose for square matrices
+ */
+ public void testTransposeInplace() {
+ if (A.isSquare())
+ assertEquals(transpose(), A.copy().transpose());
+ }
+
+ /**
+ * Scaling with an arbitrary scalar
+ */
+ public void testScale() {
+ double alpha = Math.random();
+ A = A.scale(alpha);
+ scale(alpha);
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Scaling by zero
+ */
+ public void testZeroScale() {
+ A = A.scale(0);
+ scale(0);
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Scaling by one
+ */
+ public void testOneScale() {
+ A = A.scale(1);
+ scale(1);
+ assertEquals(Ad, A);
+ }
+
+ protected double[][] scale(double alpha) {
+ for (int i = 0; i < Ad.length; ++i)
+ for (int j = 0; j < Ad[i].length; ++j)
+ Ad[i][j] *= alpha;
+ return Ad;
+ }
+
+ /**
+ * Checks the 1 norm
+ */
+ public void testOneNorm() {
+ assertEquals(norm1(Ad), A.norm(Matrix.Norm.One), tol);
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Checks the Frobenius norm
+ */
+ public void testFrobeniusNorm() {
+ assertEquals(normF(Ad), A.norm(Matrix.Norm.Frobenius), tol);
+ assertEquals(Ad, A);
+ }
+
+ /**
+ * Checks the infinity norm
+ */
+ public void testInfinityNorm() {
+ assertEquals(normInf(Ad), A.norm(Matrix.Norm.Infinity), tol);
+ assertEquals(Ad, A);
+ }
+
+ protected double norm1(double[][] A) {
+ double max = 0;
+ for (int i = 0; i < A.length; ++i) {
+ double rowsum = 0;
+ for (int j = 0; j < A[i].length; ++j)
+ rowsum += Math.abs(A[i][j]);
+ max = Math.max(rowsum, max);
+ }
+ return max;
+ }
+
+ protected double normF(double[][] A) {
+ double norm = 0;
+ for (int i = 0; i < A.length; ++i)
+ for (int j = 0; j < A[i].length; ++j)
+ norm += A[i][j] * A[i][j];
+ return Math.sqrt(norm);
+ }
+
+ protected double normInf(double[][] A) {
+ if (A.length == 0)
+ return 0;
+
+ double[] columnSum = new double[A[0].length];
+ for (int i = 0; i < A.length; ++i)
+ for (int j = 0; j < A[i].length; ++j)
+ columnSum[j] += Math.abs(A[i][j]);
+
+ double max = 0;
+ for (double d : columnSum)
+ max = Math.max(max, d);
+ return max;
+ }
+
+ /**
+ * Checks for equality between the matrix and the array
+ */
+ protected void assertEquals(double[][] Ad, Matrix A) {
+ assertTrue(A != null);
+ assertTrue(Ad != null);
+ assertTrue(A.numRows() == Ad.length);
+ for (int i = 0; i < A.numRows(); ++i) {
+ assertTrue(A.numColumns() == Ad[i].length);
+ for (int j = 0; j < A.numColumns(); ++j)
+ assertEquals(Ad[i][j], A.get(i, j), 1e-12);
+ }
+ }
+
+ /**
+ * Checks for equality between two arrays
+ */
+ protected void assertEquals(double[][] Ad, double[][] Ac) {
+ assertTrue(Ac.length == Ad.length);
+ for (int i = 0; i < A.numRows(); ++i) {
+ assertTrue(Ac[i].length == Ad[i].length);
+ for (int j = 0; j < A.numColumns(); ++j)
+ assertEquals(Ad[i][j], Ac[i][j], 1e-12);
+ }
+ }
+
+ protected void assertEquals(double[] xd, Vector x) {
+ assertEquals(xd.length, x.size());
+ for (int i = 0; i < xd.length; ++i)
+ assertEquals(xd[i], x.get(i), tol);
+ }
+
+ protected void assertEquals(double[] xd, double[] yd) {
+ for (int i = 0; i < xd.length; ++i)
+ assertEquals(xd[i], yd[i], tol);
+ }
+
+ public static void assertEquals(Matrix expected, Matrix test) {
+ assertEquals(expected.numRows(), test.numRows());
+ assertEquals(expected.numColumns(), test.numColumns());
+ for (int i = 0; i < test.numRows(); i++) {
+ for (int j = 0; j < test.numColumns(); j++) {
+ assertEquals(expected.get(i, j), test.get(i, j), 0.0001);
+ }
+ }
+ }
+
+ public static void assertEquals(Vector expected, Vector test) {
+ assertEquals(expected.size(), test.size());
+ for (int i = 0; i < test.size(); i++) {
+ assertEquals(expected.get(i), test.get(i), 0.0001);
+ }
+ }
+
+ public static void assertEqualsOrOpposite(Vector expected, Vector test) {
+ assertEquals(expected.size(), test.size());
+ for (int i = 0; i < test.size(); i++) {
+ double a = expected.get(i);
+ double b = test.get(i);
+ assertTrue("abs(" + a + ") != abs(" + b + ")", Math.abs(a) - Math.abs(b) < 0.0001);
+ }
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/OrthogonalTestAbstract.java b/src/test/java/no/uib/cipr/matrix/OrthogonalTestAbstract.java
new file mode 100644
index 0000000..97b26f7
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/OrthogonalTestAbstract.java
@@ -0,0 +1,66 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import junit.framework.TestCase;
+
+/**
+ * Orthogonal matrix decomposition tests
+ */
+public abstract class OrthogonalTestAbstract extends TestCase {
+
+ /**
+ * Initial work-matrix, and non-square matrices
+ */
+ protected Matrix A, Ar, Ac;
+
+ /**
+ * Maximum matrix size, to avoid too slow tests
+ */
+ private final int max = 100;
+
+ public OrthogonalTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, n);
+ A = Matrices.random(n, n);
+ Ar = Matrices.random(n, m);
+ Ac = Matrices.random(m, n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = Ar = Ac = null;
+ }
+
+ protected void assertEquals(Matrix A, Matrix B) {
+ for (int i = 0; i < A.numRows(); ++i)
+ for (int j = 0; j < A.numColumns(); ++j)
+ assertEquals(A.get(i, j), B.get(i, j), 1e-12);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/PackCholeskyTest.java b/src/test/java/no/uib/cipr/matrix/PackCholeskyTest.java
new file mode 100644
index 0000000..98430ec
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/PackCholeskyTest.java
@@ -0,0 +1,125 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.LowerSPDPackMatrix;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.PackCholesky;
+import no.uib.cipr.matrix.UpperSPDPackMatrix;
+
+/**
+ * Tests the packed Cholesky decomposition
+ */
+public class PackCholeskyTest extends TestCase {
+
+ private LowerSPDPackMatrix L;
+
+ private UpperSPDPackMatrix U;
+
+ private DenseMatrix I;
+
+ private final int max = 50;
+
+ public PackCholeskyTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+
+ L = new LowerSPDPackMatrix(n);
+ Utilities.lowerPopulate(L);
+ Utilities.addDiagonal(L, 1);
+ while (!Utilities.spd(L))
+ Utilities.addDiagonal(L, 1);
+
+ U = new UpperSPDPackMatrix(n);
+ Utilities.upperPopulate(U);
+ Utilities.addDiagonal(U, 1);
+ while (!Utilities.spd(U))
+ Utilities.addDiagonal(U, 1);
+
+ I = Matrices.identity(n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ L = null;
+ U = null;
+ I = null;
+ }
+
+ public void testLowerPackCholesky() {
+ int n = L.numRows();
+
+ PackCholesky c = new PackCholesky(n, false);
+ c.factor(L.copy());
+
+ c.solve(I);
+
+ Matrix J = I.mult(L, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testUpperPackCholesky() {
+ int n = U.numRows();
+
+ PackCholesky c = new PackCholesky(n, true);
+ c.factor(U.copy());
+
+ c.solve(I);
+
+ Matrix J = I.mult(U, new DenseMatrix(n, n));
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < n; ++j)
+ if (i != j)
+ assertEquals(J.get(i, j), 0, 1e-10);
+ else
+ assertEquals(J.get(i, j), 1, 1e-10);
+ }
+
+ public void testLowerPackCholeskyrcond() {
+ int n = L.numRows();
+
+ PackCholesky c = new PackCholesky(n, false);
+ c.factor(L.copy());
+
+ c.rcond(L);
+ }
+
+ public void testUpperPackCholeskyrcond() {
+ int n = U.numRows();
+
+ PackCholesky c = new PackCholesky(n, true);
+ c.factor(U.copy());
+
+ c.rcond(U);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/PermutationMatrixTest.java b/src/test/java/no/uib/cipr/matrix/PermutationMatrixTest.java
new file mode 100644
index 0000000..0a099ab
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/PermutationMatrixTest.java
@@ -0,0 +1,64 @@
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+
+/**
+ * @author Sam Halliday
+ */
+public class PermutationMatrixTest extends TestCase {
+
+ Matrix m = new DenseMatrix(new double[][]{
+ {2, -1, -2},
+ {-4, 6, 3},
+ {-4, -2, 8}
+ });
+ DenseMatrix e = new DenseMatrix(new double[][]{
+ {-4, 6, 3},
+ {-4, -2, 8},
+ {2, -1, -2}
+ });
+ DenseMatrix eI = new DenseMatrix(new double[][]{
+ {-4, -2, 8},
+ {2, -1, -2},
+ {-4, 6, 3}
+ });
+ int[] piv = new int[]{2, 3, 3};
+
+ public void testMultiply() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.mult(m, new CompRowMatrix(new DenseMatrix(m.numRows(), m.numColumns())));
+ MatrixTestAbstract.assertEquals(e, c);
+ }
+
+ public void testMultiplyTrans() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.transAmult(m, new CompRowMatrix(new DenseMatrix(m.numRows(), m.numColumns())));
+ MatrixTestAbstract.assertEquals(eI, c);
+ }
+
+ public void testMultiplyDense() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.mult(m, new DenseMatrix(m.numRows(), m.numColumns()));
+ MatrixTestAbstract.assertEquals(e, c);
+ }
+
+ public void testMultiplyTransDense() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.transAmult(m, new DenseMatrix(m.numRows(), m.numColumns()));
+ MatrixTestAbstract.assertEquals(eI, c);
+ }
+
+ public void testMultiplyCompRow() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.mult(new CompRowMatrix(m), new CompRowMatrix(new DenseMatrix(m.numRows(), m.numColumns())));
+ MatrixTestAbstract.assertEquals(e, c);
+ }
+
+ public void testMultiplyTransCompRow() {
+ Matrix p = PermutationMatrix.fromPartialPivots(piv);
+ Matrix c = p.transAmult(new CompRowMatrix(m), new CompRowMatrix(new DenseMatrix(m.numRows(), m.numColumns())));
+ MatrixTestAbstract.assertEquals(eI, c);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/QLTest.java b/src/test/java/no/uib/cipr/matrix/QLTest.java
new file mode 100644
index 0000000..6c969ac
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/QLTest.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.QL;
+
+/**
+ * QL test
+ */
+public class QLTest extends OrthogonalTestAbstract {
+
+ public QLTest(String arg0) {
+ super(arg0);
+ }
+
+ public void testStaticFactorize() {
+ assertEquals(A, QL.factorize(A));
+ }
+
+ public void testRepeatStaticFactorize() {
+ assertEquals(A, QL.factorize(A));
+ assertEquals(A, QL.factorize(A));
+ }
+
+ public void testFactor() {
+ QL ql = new QL(A.numRows(), A.numColumns());
+ assertEquals(A, ql.factor(new DenseMatrix(A)));
+ }
+
+ public void testRepeatFactor() {
+ QL ql = new QL(A.numRows(), A.numColumns());
+ ql.factor(new DenseMatrix(A));
+ assertEquals(A, ql);
+ ql.factor(new DenseMatrix(A));
+ assertEquals(A, ql);
+ }
+
+ public void testStaticFactorizeNonSquare() {
+ assertEquals(Ar, QL.factorize(Ar));
+ }
+
+ public void testRepeatStaticFactorizeNonSquare() {
+ assertEquals(Ar, QL.factorize(Ar));
+ assertEquals(Ar, QL.factorize(Ar));
+ }
+
+ public void testFactorNonSquare() {
+ QL ql = new QL(Ar.numRows(), Ar.numColumns());
+ assertEquals(Ar, ql.factor(new DenseMatrix(Ar)));
+ }
+
+ public void testRepeatFactorNonSquare() {
+ QL ql = new QL(Ar.numRows(), Ar.numColumns());
+ ql.factor(new DenseMatrix(Ar));
+ assertEquals(Ar, ql);
+ ql.factor(new DenseMatrix(Ar));
+ assertEquals(Ar, ql);
+ }
+
+ private void assertEquals(Matrix A, QL ql) {
+ assertEquals(A, ql.getQ().mult(ql.getL(), A.copy().zero()));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/QRPTest.java b/src/test/java/no/uib/cipr/matrix/QRPTest.java
new file mode 100644
index 0000000..c696012
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/QRPTest.java
@@ -0,0 +1,181 @@
+/*
+ * Copyright (C) 2006 Rafael de Pelegrini Soares
+ *
+ * MTJ additions.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+
+/**
+ * QRP test
+ */
+public class QRPTest extends TestCase {
+
+ public void testIdentity() {
+ Matrix A = Matrices.identity(5);
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(A, mult(qrp.getQ(), qrp.getPVector(), qrp.getR(), A.copy().zero()));
+ }
+
+ public void testRectangularIdentity1() {
+ Matrix A = new DenseMatrix(5, 6);
+ for (MatrixEntry i : A) {
+ if (i.row() == i.column())
+ i.set(1);
+ }
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(A, mult(qrp.getQ(), qrp.getPVector(), qrp.getR(), A.copy().zero()));
+ }
+
+ public void testRectangularIdentity2() {
+ Matrix A = new DenseMatrix(6, 5);
+ for (MatrixEntry i : A) {
+ if (i.row() == i.column())
+ i.set(1);
+ }
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(A, mult(qrp.getQ(), qrp.getPVector(), qrp.getR(), A.copy().zero()));
+ }
+
+ public void testOrthogonality() {
+ Matrix A = Matrices.random(6, 4);
+
+ QRP qrp = QRP.factorize(A);
+
+ Matrix QP = qrp.getQ();
+
+ assertEquals(Matrices.identity(QP.numRows()), QP.transAmult(QP, QP.copy().zero()));
+ }
+
+ public void testOrthogonality2() {
+ Matrix A = Matrices.random(4, 6);
+
+ QRP qrp = QRP.factorize(A);
+
+ Matrix QP = qrp.getQ();
+
+ assertEquals(Matrices.identity(QP.numRows()), QP.transAmult(QP, QP.copy().zero()));
+ }
+
+ public void testRandSquare() {
+ Matrix A = Matrices.random(5, 5);
+
+ QRP qrp = QRP.factorize(A);
+ Matrix Q = qrp.getQ();
+ Matrix R = qrp.getR();
+ int P[] = qrp.getPVector();
+
+ assertEquals(A, mult(Q, P, R, A.copy().zero()));
+ }
+
+ public void testRandRectangular1() {
+ Matrix A = Matrices.random(4, 6);
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(A, mult(qrp.getQ(), qrp.getPVector(), qrp.getR(), A.copy().zero()));
+ }
+
+ public void testRandRectangular2() {
+ Matrix A = Matrices.random(6, 4);
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(A, mult(qrp.getQ(), qrp.getPVector(), qrp.getR(), A.copy().zero()));
+ }
+
+ public void testPivotingMatrix() {
+ Matrix A = Matrices.random(6, 4);
+
+ QRP qrp = QRP.factorize(A);
+ Matrix Q = qrp.getQ();
+ Matrix R = qrp.getR();
+ Matrix P = qrp.getP();
+
+ Matrix C = A.copy().zero();
+ Matrix D = A.copy().zero();
+
+ Q.mult(R, C);
+ C.transBmult(P, D);
+
+ assertEquals(A, D);
+ }
+
+ public void testRank1() {
+ Matrix rand = Matrices.random(6, 4);
+
+ Matrix A = new DenseMatrix(rand.numRows(), rand.numRows());
+ rand.transBmult(rand, A);
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(Math.min(rand.numRows(), rand.numColumns()), qrp.getRank());
+ }
+
+ public void testRank2() {
+ Matrix rand = Matrices.random(4, 6);
+
+ Matrix A = new DenseMatrix(rand.numRows(), rand.numRows());
+ rand.transBmult(rand, A);
+
+ QRP qrp = QRP.factorize(A);
+
+ assertEquals(Math.min(rand.numRows(), rand.numColumns()), qrp.getRank());
+ }
+
+ /**
+ * Executes the multiplication C = Q*P*R
+ * @param Q matrix Q
+ * @param P column permutation of R
+ * @param R matrix R
+ * @param C matrix tu put the results
+ * @return the matrix C
+ */
+ protected Matrix mult(Matrix Q, int[] P, Matrix R, Matrix C) {
+ for (int i = 0; i < Q.numRows(); ++i) {
+ for (int j = 0; j < C.numColumns(); ++j) {
+ double dot = 0;
+ for (int k = 0; k < Q.numColumns(); ++k) {
+ dot += Q.get(i, k) * R.get(k, j);
+ }
+ C.add(i, P[j], dot);
+ }
+ }
+ return C;
+ }
+ private static final double DELTA = 1e-12;
+
+ /**
+ * Assert that the given matrices are identical
+ */
+ protected void assertEquals(Matrix A, Matrix B) {
+ assertEquals(A.numRows(), B.numRows());
+ assertEquals(A.numColumns(), B.numColumns());
+ for (int i = 0; i < A.numRows(); ++i) {
+ for (int j = 0; j < A.numColumns(); ++j) {
+ assertEquals(A.get(i, j), B.get(i, j), DELTA);
+ }
+ }
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/QRTest.java b/src/test/java/no/uib/cipr/matrix/QRTest.java
new file mode 100644
index 0000000..25b2a1b
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/QRTest.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.QR;
+
+/**
+ * QR test
+ */
+public class QRTest extends OrthogonalTestAbstract {
+
+ public QRTest(String arg0) {
+ super(arg0);
+ }
+
+ public void testStaticFactorize() {
+ assertEquals(A, QR.factorize(A));
+ }
+
+ public void testRepeatStaticFactorize() {
+ assertEquals(A, QR.factorize(A));
+ assertEquals(A, QR.factorize(A));
+ }
+
+ public void testFactor() {
+ QR qr = new QR(A.numRows(), A.numColumns());
+ assertEquals(A, qr.factor(new DenseMatrix(A)));
+ }
+
+ public void testRepeatFactor() {
+ QR qr = new QR(A.numRows(), A.numColumns());
+ qr.factor(new DenseMatrix(A));
+ assertEquals(A, qr);
+ qr.factor(new DenseMatrix(A));
+ assertEquals(A, qr);
+ }
+
+ public void testStaticFactorizeNonSquare() {
+ assertEquals(Ar, QR.factorize(Ar));
+ }
+
+ public void testRepeatStaticFactorizeNonSquare() {
+ assertEquals(Ar, QR.factorize(Ar));
+ assertEquals(Ar, QR.factorize(Ar));
+ }
+
+ public void testFactorNonSquare() {
+ QR qr = new QR(Ar.numRows(), Ar.numColumns());
+ assertEquals(Ar, qr.factor(new DenseMatrix(Ar)));
+ }
+
+ public void testRepeatFactorNonSquare() {
+ QR qr = new QR(Ar.numRows(), Ar.numColumns());
+ qr.factor(new DenseMatrix(Ar));
+ assertEquals(Ar, qr);
+ qr.factor(new DenseMatrix(Ar));
+ assertEquals(Ar, qr);
+ }
+
+ private void assertEquals(Matrix A, QR qr) {
+ assertEquals(A, qr.getQ().mult(qr.getR(), A.copy().zero()));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/RQTest.java b/src/test/java/no/uib/cipr/matrix/RQTest.java
new file mode 100644
index 0000000..0afe9d4
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/RQTest.java
@@ -0,0 +1,84 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.RQ;
+
+/**
+ * RQ test
+ */
+public class RQTest extends OrthogonalTestAbstract {
+
+ public RQTest(String arg0) {
+ super(arg0);
+ }
+
+ public void testStaticFactorize() {
+ assertEquals(A, RQ.factorize(A));
+ }
+
+ public void testRepeatStaticFactorize() {
+ assertEquals(A, RQ.factorize(A));
+ assertEquals(A, RQ.factorize(A));
+ }
+
+ public void testFactor() {
+ RQ c = new RQ(A.numRows(), A.numColumns());
+ assertEquals(A, c.factor(new DenseMatrix(A)));
+ }
+
+ public void testRepeatFactor() {
+ RQ rq = new RQ(A.numRows(), A.numColumns());
+ rq.factor(new DenseMatrix(A));
+ assertEquals(A, rq);
+ rq.factor(new DenseMatrix(A));
+ assertEquals(A, rq);
+ }
+
+ public void testStaticFactorizeNonSquare() {
+ assertEquals(Ac, RQ.factorize(Ac));
+ }
+
+ public void testRepeatStaticFactorizeNonSquare() {
+ assertEquals(Ac, RQ.factorize(Ac));
+ assertEquals(Ac, RQ.factorize(Ac));
+ }
+
+ public void testFactorNonSquare() {
+ RQ rq = new RQ(Ac.numRows(), Ac.numColumns());
+ assertEquals(Ac, rq.factor(new DenseMatrix(Ac)));
+ }
+
+ public void testRepeatFactorNonSquare() {
+ RQ rq = new RQ(Ac.numRows(), Ac.numColumns());
+ rq.factor(new DenseMatrix(Ac));
+ assertEquals(Ac, rq);
+ rq.factor(new DenseMatrix(Ac));
+ assertEquals(Ac, rq);
+ }
+
+ private void assertEquals(Matrix A, RQ rq) {
+ assertEquals(A, rq.getR().mult(rq.getQ(), A.copy().zero()));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SPDTridiagMatrixTest.java b/src/test/java/no/uib/cipr/matrix/SPDTridiagMatrixTest.java
new file mode 100644
index 0000000..e85d2af
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SPDTridiagMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.SPDTridiagMatrix;
+
+/**
+ * Test of symmetrical, positive definite tridiagonal matrices
+ */
+public class SPDTridiagMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public SPDTridiagMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new SPDTridiagMatrix(n);
+ Ad = Utilities.bandPopulate(A, 1, 1);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SingularvalueTest.java b/src/test/java/no/uib/cipr/matrix/SingularvalueTest.java
new file mode 100644
index 0000000..407ad3e
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SingularvalueTest.java
@@ -0,0 +1,85 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.SVD;
+import no.uib.cipr.matrix.TridiagMatrix;
+import junit.framework.TestCase;
+
+/**
+ * Test the singular value solver
+ */
+public class SingularvalueTest extends TestCase {
+
+ /**
+ * Matrix to decompose
+ */
+ private DenseMatrix A;
+
+ /**
+ * Maximum matrix size, to avoid too slow tests
+ */
+ private final int max = 100;
+
+ public SingularvalueTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new DenseMatrix(n, n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = null;
+ }
+
+ public void testStaticFactorize() throws NotConvergedException {
+ assertEquals(A, SVD.factorize(A));
+ }
+
+ public void testFactor() throws NotConvergedException {
+ SVD svd = new SVD(A.numRows(), A.numColumns());
+ assertEquals(A, svd.factor(A.copy()));
+ }
+
+ private void assertEquals(Matrix A, SVD svd) {
+ TridiagMatrix S = new TridiagMatrix(svd.getS().length);
+ System.arraycopy(svd.getS(), 0, S.getDiagonal(), 0, svd.getS().length);
+ DenseMatrix U = svd.getU();
+ DenseMatrix Vt = svd.getVt();
+
+ // Compute U*S*Vt
+ Matrix s = U.mult(S.mult(Vt, new DenseMatrix(S.numRows(), Vt
+ .numColumns())), new DenseMatrix(A.numRows(), A.numColumns()));
+
+ // Check that A=U*S*Vt
+ for (int i = 0; i < A.numRows(); ++i)
+ for (int j = 0; j < A.numColumns(); ++j)
+ assertEquals(A.get(i, j), s.get(i, j), 1e-12);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SquareDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/SquareDenseMatrixTest.java
new file mode 100644
index 0000000..43184c5
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SquareDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+
+/**
+ * Test of square dense matrices. This is done as some solvers change for square
+ * matrices (LU and QR)
+ */
+public class SquareDenseMatrixTest extends MatrixTestAbstract {
+
+ public SquareDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new DenseMatrix(n, n);
+ Ad = Utilities.populate(A);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/StructImmutableMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/StructImmutableMatrixTestAbstract.java
new file mode 100644
index 0000000..4b2b016
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/StructImmutableMatrixTestAbstract.java
@@ -0,0 +1,142 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Test of a matrix whose structure cannot change, but its numerical values can
+ */
+public abstract class StructImmutableMatrixTestAbstract extends MatrixTestAbstract {
+
+ public StructImmutableMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ public void testMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testOneMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testOneMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testRandomMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testRandomMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testZeroMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testZeroMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testVectorRank1() {
+ // Not supported
+ }
+
+ @Override
+ public void testVectorRank1Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testVectorRank2() {
+ // Not supported
+ }
+
+ @Override
+ public void testVectorRank2Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixRank1() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixRank1Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixTransRank1Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixTransRank1() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixRank2() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixRank2Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixTransRank2() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixTransRank2Dense() {
+ // Not supported
+ }
+
+ @Override
+ public void testTranspose() {
+ // Not supported
+ }
+
+ @Override
+ public void testTransposeInplace() {
+ // Not supported
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmBandEigenvalueTest.java b/src/test/java/no/uib/cipr/matrix/SymmBandEigenvalueTest.java
new file mode 100644
index 0000000..1023315
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmBandEigenvalueTest.java
@@ -0,0 +1,94 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmBandMatrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.SymmBandEVD;
+import no.uib.cipr.matrix.UpperSymmBandMatrix;
+
+/**
+ * Test of the symmetric, tridiagonal eigenvalue solver
+ */
+public class SymmBandEigenvalueTest extends SymmEigenvalueTestAbstract {
+
+ private LowerSymmBandMatrix L;
+
+ private UpperSymmBandMatrix U;
+
+ public SymmBandEigenvalueTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ super.setUp();
+ int kd = Utilities.getInt(1, A.numRows());
+ L = new LowerSymmBandMatrix(A, kd);
+ U = new UpperSymmBandMatrix(A, kd);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ super.tearDown();
+ L = null;
+ U = null;
+ }
+
+ public void testLowerStaticFactorize() throws NotConvergedException {
+ SymmBandEVD evd = SymmBandEVD.factorize(L, L.numSubDiagonals());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperStaticFactorize() throws NotConvergedException {
+ SymmBandEVD evd = SymmBandEVD.factorize(U, U.numSuperDiagonals());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerFactor() throws NotConvergedException {
+ SymmBandEVD evd = new SymmBandEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperFactor() throws NotConvergedException {
+ SymmBandEVD evd = new SymmBandEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerRepeatFactor() throws NotConvergedException {
+ SymmBandEVD evd = new SymmBandEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperRepeatFactor() throws NotConvergedException {
+ SymmBandEVD evd = new SymmBandEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmDenseEigenvalueTest.java b/src/test/java/no/uib/cipr/matrix/SymmDenseEigenvalueTest.java
new file mode 100644
index 0000000..b98a541
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmDenseEigenvalueTest.java
@@ -0,0 +1,93 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmDenseMatrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.SymmDenseEVD;
+import no.uib.cipr.matrix.UpperSymmDenseMatrix;
+
+/**
+ * Test of the symmetric, dense eigenvalue solver
+ */
+public class SymmDenseEigenvalueTest extends SymmEigenvalueTestAbstract {
+
+ private LowerSymmDenseMatrix L;
+
+ private UpperSymmDenseMatrix U;
+
+ public SymmDenseEigenvalueTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ super.setUp();
+ L = new LowerSymmDenseMatrix(A);
+ U = new UpperSymmDenseMatrix(A);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ super.tearDown();
+ L = null;
+ U = null;
+ }
+
+ public void testLowerStaticFactorize() throws NotConvergedException {
+ SymmDenseEVD evd = SymmDenseEVD.factorize(L);
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperStaticFactorize() throws NotConvergedException {
+ SymmDenseEVD evd = SymmDenseEVD.factorize(U);
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerFactor() throws NotConvergedException {
+ SymmDenseEVD evd = new SymmDenseEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperFactor() throws NotConvergedException {
+ SymmDenseEVD evd = new SymmDenseEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerRepeatFactor() throws NotConvergedException {
+ SymmDenseEVD evd = new SymmDenseEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperRepeatFactor() throws NotConvergedException {
+ SymmDenseEVD evd = new SymmDenseEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmEigenvalueTestAbstract.java b/src/test/java/no/uib/cipr/matrix/SymmEigenvalueTestAbstract.java
new file mode 100644
index 0000000..09e8b1d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmEigenvalueTestAbstract.java
@@ -0,0 +1,74 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import junit.framework.TestCase;
+
+/**
+ * Tests the symmetric eigenvalue computers
+ */
+public abstract class SymmEigenvalueTestAbstract extends TestCase {
+
+ /**
+ * Initial work-matrix
+ */
+ protected Matrix A;
+
+ /**
+ * Maximum matrix size, to avoid too slow tests
+ */
+ private final int max = 100;
+
+ public SymmEigenvalueTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(max);
+ A = Matrices.random(n, n);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = null;
+ }
+
+ protected void assertEquals(Matrix A, double[] w, DenseMatrix Z) {
+ // A*X
+ Matrix left = A.mult(Z, new DenseMatrix(A.numRows(), A.numColumns()));
+
+ // lambda*X
+ Matrix right = new DenseMatrix(Z);
+ for (int i = 0; i < w.length; ++i)
+ for (int j = 0; j < w.length; ++j)
+ right.set(i, j, w[j] * right.get(i, j));
+
+ // Check that A*X=lambda*X
+ for (int i = 0; i < A.numRows(); ++i)
+ for (int j = 0; j < A.numColumns(); ++j)
+ assertEquals(left.get(i, j), right.get(i, j), 1e-12);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmPackEigenvalueTest.java b/src/test/java/no/uib/cipr/matrix/SymmPackEigenvalueTest.java
new file mode 100644
index 0000000..48f039e
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmPackEigenvalueTest.java
@@ -0,0 +1,93 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.LowerSymmPackMatrix;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.SymmPackEVD;
+import no.uib.cipr.matrix.UpperSymmPackMatrix;
+
+/**
+ * Test of the symmetric, dense eigenvalue solver
+ */
+public class SymmPackEigenvalueTest extends SymmEigenvalueTestAbstract {
+
+ private LowerSymmPackMatrix L;
+
+ private UpperSymmPackMatrix U;
+
+ public SymmPackEigenvalueTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ super.setUp();
+ L = new LowerSymmPackMatrix(A);
+ U = new UpperSymmPackMatrix(A);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ super.tearDown();
+ L = null;
+ U = null;
+ }
+
+ public void testLowerStaticFactorize() throws NotConvergedException {
+ SymmPackEVD evd = SymmPackEVD.factorize(L);
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperStaticFactorize() throws NotConvergedException {
+ SymmPackEVD evd = SymmPackEVD.factorize(U);
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerFactor() throws NotConvergedException {
+ SymmPackEVD evd = new SymmPackEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperFactor() throws NotConvergedException {
+ SymmPackEVD evd = new SymmPackEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testLowerRepeatFactor() throws NotConvergedException {
+ SymmPackEVD evd = new SymmPackEVD(A.numRows(), false);
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(L.copy());
+ assertEquals(L, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testUpperRepeatFactor() throws NotConvergedException {
+ SymmPackEVD evd = new SymmPackEVD(A.numRows(), true);
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(U.copy());
+ assertEquals(U, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmTridiagEigenvalueTest.java b/src/test/java/no/uib/cipr/matrix/SymmTridiagEigenvalueTest.java
new file mode 100644
index 0000000..05a2297
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmTridiagEigenvalueTest.java
@@ -0,0 +1,67 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.NotConvergedException;
+import no.uib.cipr.matrix.SymmTridiagEVD;
+import no.uib.cipr.matrix.SymmTridiagMatrix;
+
+/**
+ * Test of the symmetric, tridiagonal eigenvalue solver
+ */
+public class SymmTridiagEigenvalueTest extends SymmEigenvalueTestAbstract {
+
+ private SymmTridiagMatrix T;
+
+ private final int max = 100;
+
+ public SymmTridiagEigenvalueTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = Matrices.random(n, n);
+ T = new SymmTridiagMatrix(A);
+ }
+
+ public void testStaticFactorize() throws NotConvergedException {
+ SymmTridiagEVD evd = SymmTridiagEVD.factorize(A);
+ assertEquals(T, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testFactor() throws NotConvergedException {
+ SymmTridiagEVD evd = new SymmTridiagEVD(A.numRows());
+ evd.factor(T.copy());
+ assertEquals(T, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+ public void testRepeatFactor() throws NotConvergedException {
+ SymmTridiagEVD evd = new SymmTridiagEVD(A.numRows());
+ evd.factor(T.copy());
+ assertEquals(T, evd.getEigenvalues(), evd.getEigenvectors());
+ evd.factor(T.copy());
+ assertEquals(T, evd.getEigenvalues(), evd.getEigenvectors());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmTridiagMatrixTest.java b/src/test/java/no/uib/cipr/matrix/SymmTridiagMatrixTest.java
new file mode 100644
index 0000000..8069086
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmTridiagMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.SymmTridiagMatrix;
+
+/**
+ * Test of SymmTridiagMatrix
+ */
+public class SymmTridiagMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public SymmTridiagMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new SymmTridiagMatrix(n);
+ Ad = Utilities.bandPopulate(A, 1, 1);
+ Utilities.lowerSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/SymmetricMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/SymmetricMatrixTestAbstract.java
new file mode 100644
index 0000000..e108f94
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/SymmetricMatrixTestAbstract.java
@@ -0,0 +1,79 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Test of symmetrical matrices
+ */
+public abstract class SymmetricMatrixTestAbstract extends MatrixTestAbstract {
+
+ public SymmetricMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ public void testMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testOneMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testOneMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testRandomMatrixAdd() {
+ // Not supported
+ }
+
+ @Override
+ public void testRandomMatrixSet() {
+ // Not supported
+ }
+
+ @Override
+ public void testVectorRank1() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank1(alpha, xdR, xdR), A.rank1(alpha, xR, xR));
+ }
+ }
+
+ @Override
+ public void testVectorRank1Dense() {
+ if (A.isSquare()) {
+ double alpha = Math.random();
+ assertEquals(rank1(alpha, xdR, xdR), A.rank1(alpha, xDenseR,
+ xDenseR));
+ }
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/TriangMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/TriangMatrixTestAbstract.java
new file mode 100644
index 0000000..7d74f01
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/TriangMatrixTestAbstract.java
@@ -0,0 +1,37 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Test of triangular matrices
+ */
+public abstract class TriangMatrixTestAbstract extends StructImmutableMatrixTestAbstract {
+
+ public TriangMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ public void testTransposeInplace() {
+ // Not supported
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/TridiagMatrixTest.java b/src/test/java/no/uib/cipr/matrix/TridiagMatrixTest.java
new file mode 100644
index 0000000..2e3e35b
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/TridiagMatrixTest.java
@@ -0,0 +1,51 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.TridiagMatrix;
+
+/**
+ * Test of tridiagonal matrices
+ */
+public class TridiagMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public TridiagMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new TridiagMatrix(n);
+ Ad = Utilities.bandPopulate(A, 1, 1);
+ }
+
+ @Override
+ public void testTransMatrixSolve() {
+ // Not supported
+ }
+
+ @Override
+ public void testTransVectorSolve() {
+ // Not supported
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrixTest.java
new file mode 100644
index 0000000..b84bbb8
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UnitLowerTriangBandMatrix;
+
+/**
+ * Test of UnitLowerTriangBandMatrix
+ */
+public class UnitLowerTriangBandMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitLowerTriangBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new UnitLowerTriangBandMatrix(n, kd);
+ Ad = Utilities.unitBandPopulate(A, kd, 0);
+ Utilities.unitSet(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrixTest.java
new file mode 100644
index 0000000..646109a
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangDenseMatrixTest.java
@@ -0,0 +1,59 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+/**
+ * Test of UnitLowerTriangDenseMatrix
+ */
+public class UnitLowerTriangDenseMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitLowerTriangDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UnitLowerTriangDenseMatrix(n);
+ Ad = Utilities.unitLowerPopulate(A);
+ Utilities.unitSet(Ad);
+ }
+
+ public void testMultUpper() {
+ Matrix lu = new DenseMatrix(new double[][]{
+ {-4.00, 6.00, 3.00},
+ {1.00, -8.00, 5.00},
+ {-0.50, -0.25, 0.75}
+ });
+ Matrix l = new UnitLowerTriangDenseMatrix(lu, false);
+ Matrix u = new UpperTriangDenseMatrix(lu, false);
+
+ Matrix e = new DenseMatrix(new double[][]{
+ {-4, 6, 3},
+ {-4, -2, 8},
+ {2, -1, -2}
+ });
+
+ Matrix out = l.mult(u, new DenseMatrix(3,3));
+ MatrixTestAbstract.assertEquals(e, out);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrixTest.java
new file mode 100644
index 0000000..762041f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitLowerTriangPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UnitLowerTriangPackMatrix;
+
+/**
+ * Test of UnitLowerTriangPackMatrix
+ */
+public class UnitLowerTriangPackMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitLowerTriangPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UnitLowerTriangPackMatrix(n);
+ Ad = Utilities.unitLowerPopulate(A);
+ Utilities.unitSet(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitTriangMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/UnitTriangMatrixTestAbstract.java
new file mode 100644
index 0000000..3cfb102
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitTriangMatrixTestAbstract.java
@@ -0,0 +1,96 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+
+/**
+ * Test of unit, triangular matrices
+ */
+public abstract class UnitTriangMatrixTestAbstract extends TriangMatrixTestAbstract {
+
+ public UnitTriangMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ public void testAddDiagonal() {
+ // Not applicable to unit triangular matrices
+ }
+
+ public void testAddOneDiagonal() {
+ // Not applicable to unit triangular matrices
+ }
+
+ public void testAddZeroDiagonal() {
+ // Not applicable to unit triangular matrices
+ }
+
+ @Override
+ public void testIteratorSet() {
+ double alpha = Math.random();
+ for (MatrixEntry e : A)
+ if (e.row() != e.column())
+ e.set(e.get() * alpha);
+ assertEquals(Utilities.unitSet(scale(alpha)), A);
+ }
+
+ @Override
+ public void testIteratorSetGet() {
+ // Not applicable to unit triangular matrices
+ }
+
+ @Override
+ public void testScale() {
+ // Not applicable to unit triangular matrices
+ }
+
+ @Override
+ public void testZero() {
+ // Not applicable to unit triangular matrices
+ }
+
+ @Override
+ public void testZeroScale() {
+ // Not applicable to unit triangular matrices
+ }
+
+ /**
+ * We can't zero, so we do without
+ */
+ @Override
+ public void testCopy() {
+ Matrix Ac = A.copy();
+ assertEquals(Ad, Ac);
+ }
+
+ @Override
+ public void testAdd() {
+ double alpha = Math.random();
+ for (MatrixEntry e : A)
+ if (e.row() != e.column()) {
+ A.add(e.row(), e.column(), alpha);
+ A.add(e.row(), e.column(), -alpha);
+ }
+ assertEquals(Ad, A);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrixTest.java
new file mode 100644
index 0000000..86061e6
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UnitUpperTriangBandMatrix;
+
+/**
+ * Test of UnitUpperTriangBandMatrix
+ */
+public class UnitUpperTriangBandMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitUpperTriangBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new UnitUpperTriangBandMatrix(n, kd);
+ Ad = Utilities.unitBandPopulate(A, 0, kd);
+ Utilities.unitSet(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrixTest.java
new file mode 100644
index 0000000..e0ded52
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UnitUpperTriangDenseMatrix;
+
+/**
+ * Test of UnitUpperTriangDenseMatrix
+ */
+public class UnitUpperTriangDenseMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitUpperTriangDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(max);
+ A = new UnitUpperTriangDenseMatrix(n);
+ Ad = Utilities.unitUpperPopulate(A);
+ Utilities.unitSet(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrixTest.java
new file mode 100644
index 0000000..dc01bac
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UnitUpperTriangPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UnitUpperTriangPackMatrix;
+
+/**
+ * Test of UnitUpperTriangPackMatrix
+ */
+public class UnitUpperTriangPackMatrixTest extends UnitTriangMatrixTestAbstract {
+
+ public UnitUpperTriangPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UnitUpperTriangPackMatrix(n);
+ Ad = Utilities.unitUpperPopulate(A);
+ Utilities.unitSet(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSPDBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSPDBandMatrixTest.java
new file mode 100644
index 0000000..2ce983d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSPDBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSPDBandMatrix;
+
+/**
+ * Test of UpperSPDBandMatrix
+ */
+public class UpperSPDBandMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public UpperSPDBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new UpperSPDBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, kd, kd);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSPDDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSPDDenseMatrixTest.java
new file mode 100644
index 0000000..68b10b0
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSPDDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSPDDenseMatrix;
+
+/**
+ * Test of UpperSPDDenseMatrix
+ */
+public class UpperSPDDenseMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public UpperSPDDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperSPDDenseMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSPDPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSPDPackMatrixTest.java
new file mode 100644
index 0000000..61cea07
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSPDPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSPDPackMatrix;
+
+/**
+ * Test of UpperSPDPackMatrix
+ */
+public class UpperSPDPackMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public UpperSPDPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperSPDPackMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSymmBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSymmBandMatrixTest.java
new file mode 100644
index 0000000..ac90259
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSymmBandMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSymmBandMatrix;
+
+/**
+ * Test of UpperSymmBandMatrix
+ */
+public class UpperSymmBandMatrixTest extends StructImmutableMatrixTestAbstract {
+
+ public UpperSymmBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new UpperSymmBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, kd, kd);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSymmDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSymmDenseMatrixTest.java
new file mode 100644
index 0000000..15ac202
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSymmDenseMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSymmDenseMatrix;
+
+/**
+ * Test of UpperSymmDenseMatrix
+ */
+public class UpperSymmDenseMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public UpperSymmDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperSymmDenseMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperSymmPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperSymmPackMatrixTest.java
new file mode 100644
index 0000000..6e65cb4
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperSymmPackMatrixTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperSymmPackMatrix;
+
+/**
+ * Test of UpperSymmPackMatrixTest
+ */
+public class UpperSymmPackMatrixTest extends SymmetricMatrixTestAbstract {
+
+ public UpperSymmPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperSymmPackMatrix(n);
+ Ad = Utilities.populate(A);
+ Utilities.upperSymmetrice(Ad);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperTriangBandMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperTriangBandMatrixTest.java
new file mode 100644
index 0000000..1f79f4c
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperTriangBandMatrixTest.java
@@ -0,0 +1,45 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperTriangBandMatrix;
+
+/**
+ * Test of UpperTriangBandMatrix
+ */
+public class UpperTriangBandMatrixTest extends TriangMatrixTestAbstract {
+
+ public UpperTriangBandMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int kd = Math.min(Utilities.getInt(max), Math.max(n - 1, 0));
+ A = new UpperTriangBandMatrix(n, kd);
+ Ad = Utilities.bandPopulate(A, 0, kd);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperTriangDenseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperTriangDenseMatrixTest.java
new file mode 100644
index 0000000..8b136d5
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperTriangDenseMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperTriangDenseMatrix;
+
+/**
+ * Test of UpperTriangDenseMatrix
+ */
+public class UpperTriangDenseMatrixTest extends TriangMatrixTestAbstract {
+
+ public UpperTriangDenseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperTriangDenseMatrix(n);
+ Ad = Utilities.upperPopulate(A);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/UpperTriangPackMatrixTest.java b/src/test/java/no/uib/cipr/matrix/UpperTriangPackMatrixTest.java
new file mode 100644
index 0000000..dbcd43f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/UpperTriangPackMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import no.uib.cipr.matrix.UpperTriangPackMatrix;
+
+/**
+ * Test of UpperTriangPackMatrix
+ */
+public class UpperTriangPackMatrixTest extends TriangMatrixTestAbstract {
+
+ public UpperTriangPackMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ A = new UpperTriangPackMatrix(n);
+ Ad = Utilities.upperPopulate(A);
+
+ // This ensures non-singularity
+ Utilities.addDiagonal(A, Ad, 1);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/Utilities.java b/src/test/java/no/uib/cipr/matrix/Utilities.java
new file mode 100644
index 0000000..eb39f4a
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/Utilities.java
@@ -0,0 +1,516 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import java.util.Arrays;
+import java.util.HashSet;
+import java.util.Random;
+import java.util.Set;
+
+/**
+ * Utilities for the testers
+ *
+ * NOTE: many of these random matrices are not guaranteed to have solutions
+ */
+public final class Utilities {
+
+ private Utilities() {
+ // No need to instantiate
+ }
+
+ /**
+ * Populates the sparse matrix in a symmetric fashion
+ *
+ * @param A
+ * Matrix to populate
+ * @param num
+ * Number of entries on each symmetry band
+ * @return The matrix data in dense format
+ */
+ public static double[][] symmetryPopulate(Matrix A, int num) {
+ int n = A.numRows(), m = A.numColumns();
+ if (m != n)
+ throw new IllegalArgumentException("m != n");
+ double[][] values = new double[n][m];
+ if (n == 0)
+ return values;
+
+ for (int j = 0; j < m; ++j)
+ for (int i = 0; i < num; ++i) {
+ double value = Math.random();
+ int k = (int) (Math.random() * n);
+ values[k][j] = value;
+ values[j][k] = value;
+ A.set(k, j, value);
+ A.set(j, k, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the matrix, putting a given number of entries on each column
+ *
+ * @param A
+ * Matrix to populate
+ * @param num
+ * Number of entries on each column
+ * @return The matrix data in dense format
+ */
+ public static double[][] columnPopulate(Matrix A, int num) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ if (n == 0)
+ return values;
+
+ for (int j = 0; j < m; ++j)
+ for (int i = 0; i < num; ++i) {
+ double value = Math.random();
+ int k = (int) (Math.random() * n);
+ values[k][j] = value;
+ A.set(k, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the matrix, putting a given number of entries on each row
+ *
+ * @param A
+ * Matrix to populate
+ * @param num
+ * Number of entries on each row
+ * @return The matrix data in dense format
+ */
+ public static double[][] rowPopulate(Matrix A, int num) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ if (m == 0)
+ return values;
+
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < num; ++j) {
+ double value = Math.random();
+ int k = (int) (Math.random() * m);
+ values[i][k] = value;
+ A.set(i, k, value);
+ }
+ return values;
+ }
+
+ /**
+ * Gets a row-wise non-zero pattern suitable for creating compressed row
+ * matrices
+ *
+ * @param n
+ * Number of rows
+ * @param m
+ * Number of columns
+ * @param b
+ * Number of entries on each row
+ */
+ public static int[][] getRowPattern(int n, int m, int b) {
+ int[][] nz = new int[n][];
+
+ for (int i = 0; i < n; ++i) {
+ Set<Integer> row = new HashSet<Integer>();
+ for (int j = 0; j < b; ++j)
+ row.add(getInt(m));
+
+ nz[i] = new int[row.size()];
+ int j = 0;
+ for (Integer colind : row)
+ nz[i][j++] = colind;
+ }
+
+ return nz;
+ }
+
+ /**
+ * Gets a column-wise non-zero pattern suitable for creating compressed
+ * column matrices
+ *
+ * @param n
+ * Number of rows
+ * @param m
+ * Number of columns
+ * @param b
+ * Number of entries on each column
+ */
+ public static int[][] getColumnPattern(int n, int m, int b) {
+ int[][] nz = new int[m][];
+
+ for (int i = 0; i < m; ++i) {
+ Set<Integer> column = new HashSet<Integer>();
+ for (int j = 0; j < b; ++j)
+ column.add(getInt(n));
+
+ nz[i] = new int[column.size()];
+ int j = 0;
+ for (Integer rowind : column)
+ nz[i][j++] = rowind;
+ }
+
+ return nz;
+ }
+
+ /**
+ * Populates the matrix, using the given non-zero pattern
+ *
+ * @param A
+ * Matrix to populate
+ * @param nz
+ * Column indices on each row
+ * @return The matrix data in dense format
+ */
+ public static double[][] rowPopulate(Matrix A, int[][] nz) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ if (m == 0)
+ return values;
+
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < nz[i].length; ++j) {
+ double value = Math.random();
+ int k = nz[i][j];
+ values[i][k] = value;
+ A.set(i, k, value);
+ }
+
+ return values;
+ }
+
+ /**
+ * Populates the matrix, using the given non-zero pattern
+ *
+ * @param A
+ * Matrix to populate
+ * @param nz
+ * Row indices on each column
+ * @return The matrix data in dense format
+ */
+ public static double[][] columnPopulate(Matrix A, int[][] nz) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ if (n == 0)
+ return values;
+
+ for (int j = 0; j < m; ++j)
+ for (int i = 0; i < nz[j].length; ++i) {
+ double value = Math.random();
+ int k = nz[j][i];
+ values[k][j] = value;
+ A.set(k, j, value);
+ }
+
+ return values;
+ }
+
+ /**
+ * Populates the matrix fully
+ *
+ * @param A
+ * Matrix to populate
+ * @return The matrix data in dense format
+ */
+ public static double[][] populate(Matrix A) {
+ Random random = new Random();
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j) {
+ double value = random.nextGaussian();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the banded matrix
+ *
+ * @param A
+ * Matrix to populate
+ * @param kl
+ * Number of subdiagonls
+ * @param ku
+ * Number of superdiagonals
+ * @return The matrix data in dense format
+ */
+ public static double[][] bandPopulate(Matrix A, int kl, int ku) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (j - ku <= i && i <= j + kl) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the banded matrix, but not its main diagonal
+ *
+ * @param A
+ * Matrix to populate
+ * @param kl
+ * Number of subdiagonls
+ * @param ku
+ * Number of superdiagonals
+ * @return The matrix data in dense format
+ */
+ public static double[][] unitBandPopulate(Matrix A, int kl, int ku) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (i != j && j - ku <= i && i <= j + kl) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the lower triangular part of the matrix
+ *
+ * @param A
+ * Matrix to populate
+ * @return The matrix data in dense format
+ */
+ public static double[][] lowerPopulate(Matrix A) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (j <= i) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the upper triangular part of the matrix
+ *
+ * @param A
+ * Matrix to populate
+ * @return The matrix data in dense format
+ */
+ public static double[][] upperPopulate(Matrix A) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (i <= j) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ public static void upperPopulateGauss(Matrix A) {
+ Random random = new Random();
+ for (int i = 0; i < A.numRows(); i++)
+ for (int j = 0; j <= i; j++)
+ A.set(i, j, random.nextGaussian());
+ }
+
+ /**
+ * Populates the strictly lower triangular part of the matrix
+ *
+ * @param A
+ * Matrix to populate
+ * @return The matrix data in dense format
+ */
+ public static double[][] unitLowerPopulate(Matrix A) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (j < i) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Populates the strictly upper triangular part of the matrix
+ *
+ * @param A
+ * Matrix to populate
+ * @return The matrix data in dense format
+ */
+ public static double[][] unitUpperPopulate(Matrix A) {
+ int n = A.numRows(), m = A.numColumns();
+ double[][] values = new double[n][m];
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < m; ++j)
+ if (i < j) {
+ double value = Math.random();
+ values[i][j] = value;
+ A.set(i, j, value);
+ }
+ return values;
+ }
+
+ /**
+ * Puts the upper triangular part into the lower triangle
+ */
+ public static void lowerSymmetrice(double[][] Ad) {
+ int n = Ad.length;
+ for (int i = 0; i < n; ++i)
+ for (int j = 0; j < i; ++j)
+ Ad[j][i] = Ad[i][j];
+ }
+
+ /**
+ * Puts the lower triangular part into the upper triangle
+ */
+ public static void upperSymmetrice(double[][] Ad) {
+ int n = Ad.length;
+ for (int i = 0; i < n; ++i) {
+ int m = Ad[0].length;
+ for (int j = i + 1; j < m; ++j)
+ Ad[j][i] = Ad[i][j];
+ }
+ }
+
+ /**
+ * Sets one on the main diagonal
+ */
+ public static double[][] unitSet(double[][] Ad) {
+ for (int i = 0; i < Ad.length; ++i)
+ Ad[i][i] = 1;
+ return Ad;
+ }
+
+ private final static Random r = new Random();
+
+ /**
+ * Returns an integer between zero (inclusive) and max (exclusive)
+ */
+ public static int getInt(int max) {
+ return r.nextInt(max);
+ }
+
+ /**
+ * Returns an integer between min (inclusive) and max (exclusive)
+ */
+ public static int getInt(int min, int max) {
+ return Math.max(min, getInt(max));
+ }
+
+ /**
+ * Returns true if the matrix is singular
+ */
+ public static boolean singular(Matrix A) throws NotConvergedException {
+ SVD svd = SVD.factorize(A);
+ double[] S = svd.getS();
+ for (int i = 0; i < S.length; ++i)
+ if (S[i] == 0)
+ return true;
+ return false;
+ }
+
+ /**
+ * Returns true if the matrix is positive definite
+ */
+ public static boolean spd(Matrix A) throws NotConvergedException {
+ EVD evd = EVD.factorize(A);
+ {
+ double[] S = evd.getRealEigenvalues();
+ for (int i = 0; i < S.length; ++i)
+ if (S[i] <= 0.)
+ return false;
+ }
+ {
+ double[] S = evd.getImaginaryEigenvalues();
+ for (int i = 0; i < S.length; ++i)
+ if (Math.abs(S[i]) > 1e-10)
+ return false;
+ }
+ return true;
+ }
+
+ /**
+ * Populates the given vector, and returns an array containing its values
+ */
+ public static double[] populate(Vector x) {
+ double[] xd = new double[x.size()];
+ for (int i = 0; i < x.size(); ++i) {
+ double alpha = Math.random();
+ xd[i] = alpha;
+ x.set(i, alpha);
+ }
+ return xd;
+ }
+
+ /**
+ * Populates the given vector, and returns an array containing its values.
+ * Only m entries are inserted
+ */
+ public static double[] populate(Vector x, int m) {
+ double[] xd = new double[x.size()];
+ for (int i = 0; i < m; ++i) {
+ double alpha = Math.random();
+ int k = (int) (Math.random() * x.size());
+ xd[k] = alpha;
+ x.set(k, alpha);
+ }
+ return xd;
+ }
+
+ /**
+ * Zeros the given array
+ */
+ public static void zero(double[][] A) {
+ for (int i = 0; i < A.length; ++i)
+ Arrays.fill(A[i], 0);
+ }
+
+ /**
+ * Adds to the diagonal of both the matrix and the array
+ */
+ public static void addDiagonal(Matrix A, double[][] Ad, double shift) {
+ int n = A.numRows(), m = A.numColumns();
+ for (int i = 0; i < Math.min(m, n); ++i) {
+ A.add(i, i, shift);
+ Ad[i][i] += shift;
+ }
+ }
+
+ /**
+ * Adds to the diagonal of the matrix
+ */
+ public static void addDiagonal(Matrix A, double shift) {
+ int n = A.numRows(), m = A.numColumns();
+ for (int i = 0; i < Math.min(m, n); ++i)
+ A.add(i, i, shift);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/VectorTestAbstract.java b/src/test/java/no/uib/cipr/matrix/VectorTestAbstract.java
new file mode 100644
index 0000000..630242d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/VectorTestAbstract.java
@@ -0,0 +1,296 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix;
+
+import junit.framework.TestCase;
+import no.uib.cipr.matrix.Vector.Norm;
+
+/**
+ * Test of vectors
+ */
+public abstract class VectorTestAbstract extends TestCase {
+
+ /**
+ * Max vector size
+ */
+ protected int max = 100;
+
+ /**
+ * Vector to test
+ */
+ protected Vector x;
+
+ /**
+ * Data of the vector
+ */
+ protected double[] xd;
+
+ /**
+ * Vectors of the same size as x, dense and non-dense
+ */
+ protected Vector y, yDense, z, zDense;
+
+ /**
+ * Data of those vectors
+ */
+ protected double[] yd, zd;
+
+ /**
+ * Tolerance for floating-point comparisons
+ */
+ protected double tol = 1e-5;
+
+ public VectorTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ createPrimary();
+ createAuxillerary();
+ }
+
+ protected abstract void createPrimary() throws Exception;
+
+ protected void createAuxillerary() throws Exception {
+ yDense = Matrices.random(x.size());
+ y = Matrices.synchronizedVector(yDense);
+ zDense = Matrices.random(x.size());
+ z = Matrices.synchronizedVector(zDense);
+ yd = Matrices.getArray(y);
+ zd = Matrices.getArray(z);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ x = null;
+ xd = null;
+ }
+
+ public void testSize() {
+ assertEquals(xd.length, x.size());
+ }
+
+ public void testCopy() {
+ Vector y = x.copy();
+ assertEquals(xd, y);
+ y.scale(Math.random());
+ assertEquals(xd, x);
+ }
+
+ public void testZero() {
+ assertEquals(scale(0), x.zero());
+ }
+
+ public void testScale() {
+ double alpha = Math.random();
+ assertEquals(scale(alpha), x.scale(alpha));
+ }
+
+ public void testScaleZero() {
+ assertEquals(scale(0), x.scale(0));
+ }
+
+ public void testScaleOne() {
+ assertEquals(scale(1), x.scale(1));
+ }
+
+ protected double[] scale(double alpha) {
+ for (int i = 0; i < xd.length; ++i)
+ xd[i] *= alpha;
+ return xd;
+ }
+
+ /*
+ * Test for Vector set(Vector)
+ */
+ public void testSetVectorDense() {
+ assertEquals(set(1, yd, 0, zd), x.set(yDense));
+ }
+
+ /*
+ * Test for Vector set(double, Vector)
+ */
+ public void testSetdoubleVectorDense() {
+ double alpha = Math.random();
+ assertEquals(set(alpha, yd, 0, zd), x.set(alpha, yDense));
+ }
+
+ /*
+ * Test for Vector set(Vector)
+ */
+ public void testSetVector() {
+ assertEquals(set(1, yd, 0, zd), x.set(y));
+ }
+
+ /*
+ * Test for Vector set(double, Vector)
+ */
+ public void testSetDoubleVector() {
+ double alpha = Math.random();
+ assertEquals(set(alpha, yd, 0, zd), x.set(alpha, y));
+ }
+
+ protected double[] set(double alpha, double[] yd, double beta, double[] zd) {
+ for (int i = 0; i < xd.length; ++i)
+ xd[i] = alpha * yd[i] + beta * zd[i];
+ return xd;
+ }
+
+ /*
+ * Test for Vector add(double, Vector)
+ */
+ public void testAddDoubleVectorDense() {
+ double alpha = Math.random();
+ assertEquals(add(alpha, yd), x.add(alpha, yDense));
+ }
+
+ /*
+ * Test for Vector add(Vector)
+ */
+ public void testAddVectorDense() {
+ assertEquals(add(1, yd), x.add(yDense));
+ }
+
+ /*
+ * Test for Vector add(double, Vector)
+ */
+ public void testAddDoubleVector() {
+ double alpha = Math.random();
+ assertEquals(add(alpha, yd), x.add(alpha, y));
+ }
+
+ /*
+ * Test for Vector add(Vector)
+ */
+ public void testAddVector() {
+ assertEquals(add(1, yd), x.add(y));
+ }
+
+ protected double[] add(double alpha, double[] yd) {
+ for (int i = 0; i < xd.length; ++i)
+ xd[i] += alpha * yd[i];
+ return xd;
+ }
+
+ public void testDotDense() {
+ assertEquals(dot(yd), x.dot(yDense), tol);
+ assertEquals(xd, x);
+ assertEquals(yd, yDense);
+ }
+
+ public void testDot() {
+ assertEquals(dot(yd), x.dot(y), tol);
+ assertEquals(xd, x);
+ assertEquals(yd, y);
+ }
+
+ protected double dot(double[] yd) {
+ double dot = 0;
+ for (int i = 0; i < yd.length; ++i)
+ dot += xd[i] * yd[i];
+ return dot;
+ }
+
+ public void testCardinality() {
+ assertEquals(cardinality(), Matrices.cardinality(x));
+ }
+
+ protected int cardinality() {
+ int nz = 0;
+ for (double d : xd)
+ if (d != 0)
+ nz++;
+ return nz;
+ }
+
+ public void testNorm1() {
+ assertEquals(norm1(), x.norm(Norm.One), tol);
+ }
+
+ public void testNorm2() {
+ assertEquals(norm2(), x.norm(Norm.Two), tol);
+ }
+
+ public void testNormInf() {
+ assertEquals(normInf(), x.norm(Norm.Infinity), tol);
+ }
+
+ protected double norm1() {
+ double norm = 0;
+ for (double d : xd)
+ norm += Math.abs(d);
+ return norm;
+ }
+
+ protected double norm2() {
+ double norm = 0;
+ for (double d : xd)
+ norm += d * d;
+ return Math.sqrt(norm);
+ }
+
+ protected double normInf() {
+ double norm = 0;
+ for (double d : xd)
+ norm = Math.max(Math.abs(d), norm);
+ return norm;
+ }
+
+ public void testIteratorSetGet() {
+ double alpha = Math.random();
+ double[] data = new double[x.size()];
+ for (VectorEntry e : x) {
+ data[e.index()] = e.get();
+ e.set(alpha * e.get());
+ e.set(e.get() / alpha);
+ }
+ assertEquals(xd, x);
+ assertEquals(xd, data);
+ }
+
+ public void testIteratorGet() {
+ double[] data = new double[x.size()];
+ for (VectorEntry e : x)
+ data[e.index()] = e.get();
+ assertEquals(xd, x);
+ assertEquals(xd, data);
+ }
+
+ public void testIteratorSet() {
+ double alpha = Math.random();
+ for (VectorEntry e : x)
+ e.set(alpha * e.get());
+ assertEquals(scale(alpha), x);
+ }
+
+ protected void assertEquals(double[] xd, Vector x) {
+ assertEquals(xd.length, x.size());
+ for (int i = 0; i < xd.length; ++i)
+ assertEquals(xd[i], x.get(i), tol);
+ }
+
+ protected void assertEquals(double[] xd, double[] yd) {
+ for (int i = 0; i < xd.length; ++i)
+ assertEquals(xd[i], yd[i], tol);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/io/MatrixVectorIoTest.java b/src/test/java/no/uib/cipr/matrix/io/MatrixVectorIoTest.java
new file mode 100644
index 0000000..e65974f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/io/MatrixVectorIoTest.java
@@ -0,0 +1,28 @@
+package no.uib.cipr.matrix.io;
+
+import junit.framework.TestCase;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixTestAbstract;
+
+import java.io.BufferedWriter;
+import java.io.File;
+import java.io.FileReader;
+import java.io.FileWriter;
+
+public class MatrixVectorIoTest extends TestCase {
+
+ public void testWriteRead() throws Exception {
+ DenseMatrix mat = new DenseMatrix(new double[][] {{1.1,1.2},{1.3,1.4}});
+ File matrixFile = new File("TestMatrixFile");
+ BufferedWriter out = new BufferedWriter(new FileWriter(matrixFile));
+ MatrixVectorWriter writer = new MatrixVectorWriter(out);
+ writer.printMatrixInfo(new MatrixInfo(false, MatrixInfo.MatrixField.Real, MatrixInfo.MatrixSymmetry.General));
+ writer.printMatrixSize(new MatrixSize(mat.numRows(), mat.numColumns(), mat.numColumns() * mat.numRows()));
+ writer.printArray(mat.getData());
+ writer.close();
+ Matrix newMat = new DenseMatrix(new MatrixVectorReader(new FileReader(matrixFile)));
+
+ MatrixTestAbstract.assertEquals(mat, newMat);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ArpackSymTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ArpackSymTest.java
new file mode 100644
index 0000000..b730e49
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ArpackSymTest.java
@@ -0,0 +1,76 @@
+package no.uib.cipr.matrix.sparse;
+
+import junit.framework.Assert;
+import junit.framework.TestCase;
+import lombok.extern.java.Log;
+import no.uib.cipr.matrix.*;
+import no.uib.cipr.matrix.io.MatrixVectorReader;
+
+import java.io.File;
+import java.io.FileReader;
+import java.util.HashMap;
+import java.util.Map;
+
+/**
+ * @author Sam Halliday
+ */
+ at Log
+public class ArpackSymTest extends TestCase {
+
+ public void testRandomEigensystem() throws NotConvergedException {
+ for (int i = 100; i <= 500; i = i + 100) {
+ UpperSymmDenseMatrix matrix = new UpperSymmDenseMatrix(i);
+ Utilities.upperPopulateGauss(matrix);
+ Map<Double, DenseVector> evd = evdSolve(matrix);
+
+ ArpackSym solver = new ArpackSym(matrix);
+ int todo = i / 10;
+
+ Map<Double, DenseVectorSub> results = solver.solve(todo, ArpackSym.Ritz.LA);
+ Assert.assertEquals(todo, results.size());
+ for (Map.Entry<Double, DenseVectorSub> e : results.entrySet()) {
+ // exact match of eigenvector / eigenvalue is not important for random matrices
+ // as the eigenvectors should always be the Euclidean directions
+ boolean value = false, vector = false;
+ for (Map.Entry<Double, DenseVector> evdEntry : evd.entrySet()) {
+ if (e.getKey() - evdEntry.getKey() < 0.0001)
+ value = true;
+ if (e.getValue().dot(evdEntry.getValue()) < 0.9999)
+ vector = true;
+ }
+ if (!value)
+ fail("no matching eigenvalue for " + e.getKey() + " : " + evd.keySet());
+ if (!vector)
+ fail("no matching eigenvector for " + e.getKey() + " : " + evd.keySet());
+ }
+ }
+ }
+
+ private Map<Double, DenseVector> evdSolve(UpperSymmDenseMatrix matrix) throws NotConvergedException {
+ SymmDenseEVD evd = new SymmDenseEVD(matrix.numColumns(), true);
+ evd.factor(matrix);
+ Map<Double, DenseVector> results = new HashMap<Double, DenseVector>();
+
+ double[] eigenvalues = evd.getEigenvalues();
+ Matrix eigenvectorMatrix = evd.getEigenvectors();
+ for (int i = 0; i < eigenvalues.length; i++) {
+ double eigenvalue = eigenvalues[i];
+ DenseVector eigenvector = Matrices.getColumn(eigenvectorMatrix, i);
+ results.put(eigenvalue, eigenvector);
+ }
+ return results;
+ }
+
+ public static void main(String[] args) throws Exception {
+ File file = new File("A.txt");
+ Matrix A = new LinkedSparseMatrix(new MatrixVectorReader(new FileReader(file)));
+ Matrix AtA = A.transAmult(A, new LinkedSparseMatrix(A.numColumns(), A.numColumns()));
+
+ ArpackSym solver = new ArpackSym(AtA);
+ Map<Double, DenseVectorSub> results = solver.solve(A.numRows() / 10, ArpackSym.Ritz.LA);
+ for (Map.Entry<Double, DenseVectorSub> result : results.entrySet()) {
+ log.info(result.getKey().toString());
+ }
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGDiagonalTest.java
new file mode 100644
index 0000000..8e6f92a
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of BiCG with diagonal preconditioning
+ */
+public class BiCGDiagonalTest extends BiCGTest {
+
+ public BiCGDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGILUTest.java
new file mode 100644
index 0000000..d6cee68
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGILUTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of BiCG with ILU
+ */
+public class BiCGILUTest extends BiCGTest {
+
+ public BiCGILUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILU(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGTest.java
new file mode 100644
index 0000000..851ab43
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.BiCG;
+
+/**
+ * Test of BiCG
+ */
+public class BiCGTest extends IterativeSolverTestAbstract {
+
+ public BiCGTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new BiCG(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabDiagonalTest.java
new file mode 100644
index 0000000..308dab4
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of BiCGstab with diagonal preconditioning
+ */
+public class BiCGstabDiagonalTest extends BiCGstabTest {
+
+ public BiCGstabDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTTest.java
new file mode 100644
index 0000000..313cb06
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILUT;
+
+/**
+ * Test of BiCGstab with ILUT
+ */
+public class BiCGstabILUTTest extends BiCGstabTest {
+
+ public BiCGstabILUTTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILUT(new FlexCompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTest.java
new file mode 100644
index 0000000..81db7d0
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabILUTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of BiCGstab with ILU
+ */
+public class BiCGstabILUTest extends BiCGstabTest {
+
+ public BiCGstabILUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILU(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabTest.java b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabTest.java
new file mode 100644
index 0000000..469aead
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/BiCGstabTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.BiCGstab;
+
+/**
+ * Test of BiCGstab
+ */
+public class BiCGstabTest extends IterativeSolverTestAbstract {
+
+ public BiCGstabTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new BiCGstab(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGAMGTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGAMGTest.java
new file mode 100644
index 0000000..3065905
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGAMGTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.AMG;
+
+/**
+ * Test of CG with AMG
+ */
+public class CGAMGTest extends CGTest {
+
+ public CGAMGTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new AMG();
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGDiagonalTest.java
new file mode 100644
index 0000000..17ed209
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of CG with diagonal preconditioning
+ */
+public class CGDiagonalTest extends CGTest {
+
+ public CGDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGICCTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGICCTest.java
new file mode 100644
index 0000000..bdb3f93
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGICCTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ICC;
+
+/**
+ * Test of CG with ICC
+ */
+public class CGICCTest extends CGTest {
+
+ public CGICCTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ICC(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGSDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGSDiagonalTest.java
new file mode 100644
index 0000000..fa3c687
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGSDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of CGS with diagonal preconditioning
+ */
+public class CGSDiagonalTest extends CGSTest {
+
+ public CGSDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTTest.java
new file mode 100644
index 0000000..ed4af6d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILUT;
+
+/**
+ * Test of CGS with ILUT
+ */
+public class CGSILUTTest extends CGSTest {
+
+ public CGSILUTTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILUT(new FlexCompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTest.java
new file mode 100644
index 0000000..935bca4
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGSILUTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of CGS with ILU
+ */
+public class CGSILUTest extends CGSTest {
+
+ public CGSILUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILU(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGSSORTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGSSORTest.java
new file mode 100644
index 0000000..379beca
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGSSORTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.SSOR;
+
+/**
+ * Test of CG with SSOR
+ */
+public class CGSSORTest extends CGTest {
+
+ public CGSSORTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ double omega = Math.random() + 1;
+ M = new SSOR(new CompRowMatrix(A), true, omega, omega);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGSTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGSTest.java
new file mode 100644
index 0000000..ea00150
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGSTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CGS;
+
+/**
+ * Test of CGS
+ */
+public class CGSTest extends IterativeSolverTestAbstract {
+
+ public CGSTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new CGS(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CGTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CGTest.java
new file mode 100644
index 0000000..4730a82
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CGTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CG;
+
+/**
+ * Test of CG
+ */
+public class CGTest extends SPDIterativeSolverTestAbstract {
+
+ public CGTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new CG(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevAMGTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevAMGTest.java
new file mode 100644
index 0000000..dcb2c5d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevAMGTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.AMG;
+
+/**
+ * Test of Chebyshev with AMG
+ */
+public class ChebyshevAMGTest extends ChebyshevTest {
+
+ public ChebyshevAMGTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new AMG();
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevDiagonalTest.java
new file mode 100644
index 0000000..ee9631b
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of Chebyshev with diagonal preconditioning
+ */
+public class ChebyshevDiagonalTest extends ChebyshevTest {
+
+ public ChebyshevDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevICCTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevICCTest.java
new file mode 100644
index 0000000..09614df
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevICCTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ICC;
+
+/**
+ * Test of Chebyshev with ICC
+ */
+public class ChebyshevICCTest extends ChebyshevTest {
+
+ public ChebyshevICCTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ICC(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevSSORTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevSSORTest.java
new file mode 100644
index 0000000..3f1fe14
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevSSORTest.java
@@ -0,0 +1,42 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.SSOR;
+
+/**
+ * Test of Chebyshev with SSOR preconditioning
+ */
+public class ChebyshevSSORTest extends ChebyshevTest {
+
+ public ChebyshevSSORTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ double omega = Math.random() + 1;
+ M = new SSOR(new CompRowMatrix(A), true, omega, omega);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevTest.java
new file mode 100644
index 0000000..d06331e
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ChebyshevTest.java
@@ -0,0 +1,50 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.SymmDenseEVD;
+import no.uib.cipr.matrix.sparse.Chebyshev;
+
+/**
+ * Test of Chebyshev solver
+ */
+public class ChebyshevTest extends SPDIterativeSolverTestAbstract {
+
+ public ChebyshevTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ // Get the extremal eigenvalues
+ SymmDenseEVD evd = SymmDenseEVD.factorize(A);
+ double[] eigs = evd.getEigenvalues();
+
+ double eigmin = 1, eigmax = 1;
+ if (eigs.length > 0) {
+ eigmin = eigs[0];
+ eigmax = eigs[eigs.length - 1];
+ }
+
+ solver = new Chebyshev(x, eigmin, eigmax);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CompColMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CompColMatrixTest.java
new file mode 100644
index 0000000..146cafb
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CompColMatrixTest.java
@@ -0,0 +1,45 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompColMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of CompColMatrix
+ */
+public class CompColMatrixTest extends SparseStructImmutableMatrixTestAbstract {
+
+ public CompColMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ int[][] nz = Utilities.getColumnPattern(n, m, b);
+ A = new CompColMatrix(n, m, nz);
+ Ad = Utilities.columnPopulate(A, nz);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CompDiagMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CompDiagMatrixTest.java
new file mode 100644
index 0000000..49b5477
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CompDiagMatrixTest.java
@@ -0,0 +1,43 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompDiagMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of CompDiagMatrix
+ */
+public class CompDiagMatrixTest extends SparseMatrixTestAbstract {
+
+ public CompDiagMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ A = new CompDiagMatrix(n, m);
+ Ad = Utilities.rowPopulate(A, b);
+ }
+
+}
\ No newline at end of file
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/CompRowMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/CompRowMatrixTest.java
new file mode 100644
index 0000000..edb3b6d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/CompRowMatrixTest.java
@@ -0,0 +1,45 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of CompRowMatrix
+ */
+public class CompRowMatrixTest extends SparseStructImmutableMatrixTestAbstract {
+
+ public CompRowMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ int[][] nz = Utilities.getRowPattern(n, m, b);
+ A = new CompRowMatrix(n, m, nz);
+ Ad = Utilities.rowPopulate(A, nz);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/FlexCompColMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/FlexCompColMatrixTest.java
new file mode 100644
index 0000000..3a410c5
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/FlexCompColMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompColMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of FlexCompColMatrix
+ */
+public class FlexCompColMatrixTest extends SparseMatrixTestAbstract {
+
+ public FlexCompColMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ A = new FlexCompColMatrix(n, m);
+ Ad = Utilities.columnPopulate(A, b);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrixTest.java
new file mode 100644
index 0000000..40a303b
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/FlexCompRowMatrixTest.java
@@ -0,0 +1,44 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of FlexCompRowMatrix
+ */
+public class FlexCompRowMatrixTest extends SparseMatrixTestAbstract {
+
+ public FlexCompRowMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ A = new FlexCompRowMatrix(n, m);
+ Ad = Utilities.rowPopulate(A, b);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/GMRESDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/GMRESDiagonalTest.java
new file mode 100644
index 0000000..1ee2c4f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/GMRESDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test of GMRES with diagonal preconditioning
+ */
+public class GMRESDiagonalTest extends GMRESTest {
+
+ public GMRESDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTTest.java b/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTTest.java
new file mode 100644
index 0000000..855b6f0
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILUT;
+
+/**
+ * Test of GMRES with ILUT
+ */
+public class GMRESILUTTest extends GMRESTest {
+
+ public GMRESILUTTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILUT(new FlexCompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTest.java
new file mode 100644
index 0000000..819a79f
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/GMRESILUTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of GMRES with ILU
+ */
+public class GMRESILUTest extends GMRESTest {
+
+ public GMRESILUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILU(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/GMRESTest.java b/src/test/java/no/uib/cipr/matrix/sparse/GMRESTest.java
new file mode 100644
index 0000000..69a83e2
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/GMRESTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.GMRES;
+
+/**
+ * Test of GMRES
+ */
+public class GMRESTest extends IterativeSolverTestAbstract {
+
+ public GMRESTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new GMRES(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ICCTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ICCTest.java
new file mode 100644
index 0000000..0a621af
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ICCTest.java
@@ -0,0 +1,46 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ICC;
+
+/**
+ * Test of ICC(0)
+ */
+public class ICCTest extends IncompleteFactorizationTestAbstract {
+
+ @Override
+ void testFactorization(Matrix A, Vector x) {
+ Vector b = A.mult(x, x.copy());
+
+ ICC icc = new ICC(new CompRowMatrix(A));
+ icc.setMatrix(A);
+ icc.apply(b, x);
+
+ Vector r = A.multAdd(-1, x, b.copy());
+
+ assertEquals(0, r.norm(Vector.Norm.TwoRobust), 1e-5);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ILUTTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ILUTTest.java
new file mode 100644
index 0000000..e5f144c
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ILUTTest.java
@@ -0,0 +1,46 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of ILU with thresholding
+ */
+public class ILUTTest extends IncompleteFactorizationTestAbstract {
+
+ @Override
+ void testFactorization(Matrix A, Vector x) {
+ Vector b = A.mult(x, x.copy());
+
+ ILU ilut = new ILU(new CompRowMatrix(A));
+ ilut.setMatrix(A);
+ ilut.apply(b, x);
+
+ Vector r = A.multAdd(-1, x, b.copy());
+
+ assertEquals(0, r.norm(Vector.Norm.TwoRobust), 1e-5);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/ILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/ILUTest.java
new file mode 100644
index 0000000..2461126
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/ILUTest.java
@@ -0,0 +1,45 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of ILU(0)
+ */
+public class ILUTest extends IncompleteFactorizationTestAbstract {
+
+ @Override
+ void testFactorization(Matrix A, Vector x) {
+ Vector b = A.mult(x, x.copy());
+
+ ILU ilu = new ILU(new CompRowMatrix(A));
+ ilu.setMatrix(A);
+ ilu.apply(b, x);
+
+ Vector r = A.multAdd(-1, x, b.copy());
+
+ assertEquals(0, r.norm(Vector.Norm.TwoRobust), 1e-5);
+ }
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/IncompleteFactorizationTestAbstract.java b/src/test/java/no/uib/cipr/matrix/sparse/IncompleteFactorizationTestAbstract.java
new file mode 100644
index 0000000..719da1d
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/IncompleteFactorizationTestAbstract.java
@@ -0,0 +1,79 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Utilities;
+import junit.framework.TestCase;
+
+/**
+ * Test of incomplete factorizations
+ */
+public abstract class IncompleteFactorizationTestAbstract extends TestCase {
+
+ @Override
+ protected void setUp() throws Exception {
+ super.setUp();
+ }
+
+ public void testTriDiagonal() {
+ int n = Utilities.getInt(1, 10);
+ Matrix A = new DenseMatrix(n, n);
+ Vector x = new DenseVector(n);
+
+ for (int i = 0; i < n; ++i) {
+ A.set(i, i, 2);
+ x.set(i, 1);
+ }
+ for (int i = 0; i < n - 1; ++i) {
+ A.set(i, i + 1, -1);
+ A.set(i + 1, i, -1);
+ }
+
+ testFactorization(A, x);
+ }
+
+ public void testPentaDiagonal() {
+ int n = Utilities.getInt(1, 10);
+ Matrix A = new DenseMatrix(n, n);
+ Vector x = new DenseVector(n);
+
+ for (int i = 0; i < n; ++i) {
+ A.set(i, i, 4);
+ x.set(i, 1);
+ }
+ for (int i = 0; i < n - 1; ++i) {
+ A.set(i, i + 1, -1);
+ A.set(i + 1, i, -1);
+ }
+ for (int i = 0; i < n - 2; ++i) {
+ A.set(i, i + 2, -1);
+ A.set(i + 2, i, -1);
+ }
+
+ testFactorization(A, x);
+ }
+
+ abstract void testFactorization(Matrix A, Vector x);
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/IterativeSolverTestAbstract.java b/src/test/java/no/uib/cipr/matrix/sparse/IterativeSolverTestAbstract.java
new file mode 100644
index 0000000..61220a2
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/IterativeSolverTestAbstract.java
@@ -0,0 +1,178 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.DenseLU;
+import no.uib.cipr.matrix.Matrices;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.sparse.IterativeSolver;
+import no.uib.cipr.matrix.sparse.IterativeSolverNotConvergedException;
+import no.uib.cipr.matrix.sparse.Preconditioner;
+import no.uib.cipr.matrix.Utilities;
+import junit.framework.TestCase;
+
+/**
+ * Test of the iterative solvers and preconditioners
+ */
+public abstract class IterativeSolverTestAbstract extends TestCase {
+
+ /**
+ * Number of times to repeat tests
+ */
+ private int repeat = 5;
+
+ /**
+ * Sizes of the system matrix
+ */
+ protected int max = 50, bmax = 10;
+
+ /**
+ * Numerical tolerance
+ */
+ protected double tol = 1e-4;
+
+ /**
+ * Diagonal shift for singularity handling
+ */
+ protected double shift = 100;
+
+ /**
+ * Square system matrix
+ */
+ protected Matrix A;
+
+ /**
+ * Right hand side, right hand for transpose system, and the solution vector
+ * in both cases
+ */
+ protected Vector b, bt, x;
+
+ /**
+ * Stores the data of x
+ */
+ protected double[] xd;
+
+ /**
+ * Iterative solver to use
+ */
+ protected IterativeSolver solver;
+
+ /**
+ * Preconditioner to use
+ */
+ protected Preconditioner M;
+
+ /**
+ * Constructor for IterativeSolverTestAbstract
+ */
+ public IterativeSolverTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void setUp() throws Exception {
+ createMatrix();
+
+ int n = A.numRows();
+ x = Matrices.random(n);
+ b = Matrices.random(n);
+ bt = Matrices.random(n);
+
+ // Create solver and preconditioner
+ createSolver();
+ if (M == null)
+ M = solver.getPreconditioner();
+ M.setMatrix(A);
+
+ // Compute the correct right hand sides
+ b = A.mult(x, b);
+ bt = A.transMult(x, bt);
+
+ // Store x for later. It is overwritten
+ xd = Matrices.getArray(x);
+
+ // Randomize the inital solution vector
+ Matrices.random(x);
+ }
+
+ protected abstract void createSolver() throws Exception;
+
+ protected void createMatrix() throws Exception {
+ // Create an arbitrary matrix
+ int n = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, n));
+ A = new FlexCompRowMatrix(n, n);
+ Utilities.rowPopulate(A, b);
+
+ // Make it non-singular
+ addDiagonal(A, shift);
+ DenseLU lu = DenseLU.factorize(A);
+ while (lu.isSingular()) {
+ addDiagonal(A, shift);
+ lu = DenseLU.factorize(A);
+ }
+ }
+
+ protected void addDiagonal(Matrix A, double shift) {
+ int n = A.numRows(), m = A.numColumns();
+ for (int i = 0; i < Math.min(n, m); ++i)
+ A.add(i, i, shift);
+ }
+
+ @Override
+ protected void tearDown() throws Exception {
+ A = null;
+ b = bt = x = null;
+ xd = null;
+ solver = null;
+ }
+
+ public void testSolve() {
+ try {
+ solver.solve(A, b, x);
+ assertSolved();
+ } catch (IterativeSolverNotConvergedException e) {
+ fail("Solver did not converge: " + e.getReason() + ". Residual="
+ + e.getResidual());
+ }
+ }
+
+ public void testRepeatSolve() {
+ try {
+ for (int i = 0; i < repeat; ++i) {
+ solver.solve(A, b, x);
+ assertSolved();
+ x = Matrices.random(A.numRows());
+ }
+ } catch (IterativeSolverNotConvergedException e) {
+ fail("Solver did not converge: " + e.getReason() + ". Residual="
+ + e.getResidual());
+ }
+ }
+
+ protected void assertSolved() {
+ for (int i = 0; i < xd.length; ++i)
+ assertEquals(xd[i], x.get(i), tol);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrixTest.java b/src/test/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrixTest.java
new file mode 100644
index 0000000..d31aa99
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/LinkedSparseMatrixTest.java
@@ -0,0 +1,214 @@
+package no.uib.cipr.matrix.sparse;
+
+import au.com.bytecode.opencsv.CSVWriter;
+import com.google.common.base.Stopwatch;
+import com.google.common.collect.Lists;
+import junit.framework.Assert;
+import lombok.Cleanup;
+import lombok.extern.java.Log;
+import no.uib.cipr.matrix.DenseMatrix;
+import no.uib.cipr.matrix.Matrix;
+import no.uib.cipr.matrix.MatrixEntry;
+import no.uib.cipr.matrix.Utilities;
+
+import java.io.File;
+import java.io.FileWriter;
+import java.util.concurrent.TimeUnit;
+
+/**
+ * @author Sam Halliday
+ */
+ at Log
+public class LinkedSparseMatrixTest extends SparseMatrixTestAbstract {
+ public LinkedSparseMatrixTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, m));
+ int[][] nz = Utilities.getRowPattern(n, m, b);
+ A = new LinkedSparseMatrix(n, m);
+ Ad = Utilities.rowPopulate(A, nz);
+
+ for (MatrixEntry e : A) {
+ int row = e.row();
+ int col = e.column();
+ double expect = Ad[row][col];
+ Assert.assertEquals(expect, e.get(), 0);
+ Assert.assertEquals(expect, A.get(row, col), 0);
+ }
+
+ for (int i = 0; i < n; i++) {
+ for (int j = 0; j < m; j++) {
+ double expect = Ad[i][j];
+ Assert.assertEquals(expect, A.get(i, j), 0);
+ }
+ }
+
+ LinkedSparseMatrix.Node head = ((LinkedSparseMatrix) A).links.head;
+ LinkedSparseMatrix.Node node = head;
+ while (node != null) {
+// log.info(node.toString());
+ Assert.assertEquals(Ad[node.row][node.col], node.val);
+ node = node.rowTail;
+ }
+ node = head;
+ while (node != null) {
+// log.info(node.toString());
+ Assert.assertEquals(Ad[node.row][node.col], node.val);
+ node = node.colTail;
+ }
+ }
+
+ public void ignoredTimedMult() {
+ Stopwatch watch = Stopwatch.createUnstarted();
+ DenseMatrix dense = new DenseMatrix(1000, 1000);
+ int[][] nz = Utilities.getRowPattern(dense.numRows(), dense.numColumns(), 100);
+ Utilities.rowPopulate(dense, nz);
+ log.info("created matrices");
+ Matrix sparse = new LinkedSparseMatrix(dense.numRows(), dense.numColumns());
+ sparse.set(dense);
+
+ for (Matrix m : Lists.newArrayList(dense, sparse)) {
+ log.info("starting " + m.getClass());
+ Matrix t = new DenseMatrix(m);
+ t.transpose();
+ Matrix o = new DenseMatrix(dense.numRows(), dense.numColumns());
+ log.info("warming up " + m.getClass() + " " + o.getClass());
+ for (int i = 0; i < 10; i++)
+ m.mult(t, o);
+ log.info("starting " + m.getClass() + " " + o.getClass());
+ watch.start();
+ for (int i = 0; i < 100; i++)
+ m.mult(t, o);
+ watch.stop();
+ log.info(m.getClass() + " " + o.getClass() + " " + watch);
+ }
+ }
+
+ public void ignoredTimedTransMult() {
+ Stopwatch watch = Stopwatch.createUnstarted();
+ DenseMatrix dense = new DenseMatrix(1000, 1000);
+ int[][] nz = Utilities.getRowPattern(dense.numRows(), dense.numColumns(), 100);
+ Utilities.rowPopulate(dense, nz);
+ log.info("created matrices");
+ Matrix sparse = new LinkedSparseMatrix(dense.numRows(), dense.numColumns());
+ sparse.set(dense);
+
+ for (Matrix m : Lists.newArrayList(dense, sparse)) {
+ log.info("starting " + m.getClass());
+ Matrix t = new DenseMatrix(m);
+ Matrix o = new DenseMatrix(dense.numRows(), dense.numColumns());
+ log.info("warming up " + m.getClass() + " " + o.getClass());
+ for (int i = 0; i < 10; i++)
+ m.transAmult(t, o);
+ log.info("starting " + m.getClass() + " " + o.getClass());
+ watch.start();
+ for (int i = 0; i < 100; i++)
+ m.transAmult(t, o);
+ watch.stop();
+ log.info(m.getClass() + " " + o.getClass() + " " + watch);
+ }
+ }
+
+ @Override
+ public void testIteratorSet() {
+ }
+
+ @Override
+ public void testIteratorSetGet() {
+ }
+
+
+ /**
+ * Does a naive perf test against DenseMatrix, outputting CSV
+ * that we plot in R.
+ * <p/>
+ * Generate n x n matrix with m entries, on left, and
+ * an n x n dense matrix with m entries on the right,
+ * using the same population algo. Then we multiply
+ * them and output into a dense matrix. We compare
+ * dense vs linked sparse from the left, and also
+ * look at memory usage. We repeat for 10 different
+ * values of m (10,000 to 100,000), and in both cases
+ * vary m from 1,000 to 10,000. This requires about
+ * 8GB heap to be on the safe side.
+ * <p/>
+ * -Xms8g -Xmx8g -Djava.util.logging.config.file=logging.properties
+ *
+ * @param args
+ */
+ public static void main(String[] args) throws Exception {
+ File file = new File("LinkedSparseMatrixPerf.csv");
+ log.info("writing to " + file);
+ @Cleanup CSVWriter csv = new CSVWriter(new FileWriter(file));
+
+ for (int r = 0; r < 10; r++) {
+ for (int m = 10000; m <= 100000; m = m + 10000) {
+ for (int n = 1000; n <= 10000; n = n + 1000) {
+ int[][] patternA = Utilities.getRowPattern(n, n, m / n);
+ DenseMatrix origA = new DenseMatrix(n, n);
+ Utilities.rowPopulate(origA, patternA);
+ int[][] patternB = Utilities.getRowPattern(n, n, m / n);
+ DenseMatrix origB = new DenseMatrix(n, n);
+ Utilities.rowPopulate(origB, patternB);
+ // to be fair, we reuse the same matrix values
+
+ long denseMem, denseInitTime, denseMultTime, sparseMem, sparseInitTime, sparseMultTime;
+
+ Stopwatch timer = Stopwatch.createUnstarted();
+ {
+ timer.reset();
+ timer.start();
+ DenseMatrix A = new DenseMatrix(origA);
+ timer.stop();
+ // all attempts to measure memory usage failed
+ denseMem = n * n * 8;
+ denseInitTime = timer.elapsed(TimeUnit.NANOSECONDS);
+ timer.reset();
+
+ DenseMatrix B = origB.copy();
+ DenseMatrix C = new DenseMatrix(n, n);
+ timer.start();
+ A.mult(B, C);
+ timer.stop();
+ denseMultTime = timer.elapsed(TimeUnit.NANOSECONDS);
+ }
+ {
+ timer.reset();
+ timer.start();
+ LinkedSparseMatrix A = new LinkedSparseMatrix(origA);
+ timer.stop();
+ // using compressedooms
+ sparseMem = m * 28 + 16 * n;
+ sparseInitTime = timer.elapsed(TimeUnit.NANOSECONDS);
+ timer.reset();
+
+ DenseMatrix B = origB.copy();
+ DenseMatrix C = new DenseMatrix(n, n);
+ timer.start();
+ A.mult(B, C);
+ timer.stop();
+ sparseMultTime = timer.elapsed(TimeUnit.NANOSECONDS);
+ }
+
+ String[] line = new String[]{
+ Integer.toString(n), Integer.toString(m),
+ Long.toString(denseMem), Long.toString(denseInitTime), Long.toString(denseMultTime),
+ Long.toString(sparseMem), Long.toString(sparseInitTime), Long.toString(sparseMultTime)
+ };
+ log.info(java.util.Arrays.toString(line));
+ csv.writeNext(line);
+
+ // these are to keep lots of refs above alive from GC
+ log.finest(origA.numRows() + " " + origB.numColumns() + " " + patternA.length + " " + patternB.length);
+ }
+ }
+ }
+ }
+
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/QMRDiagonalTest.java b/src/test/java/no/uib/cipr/matrix/sparse/QMRDiagonalTest.java
new file mode 100644
index 0000000..036fafd
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/QMRDiagonalTest.java
@@ -0,0 +1,40 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.DiagonalPreconditioner;
+
+/**
+ * Test QMR with diagonal preconditioning
+ */
+public class QMRDiagonalTest extends QMRTest {
+
+ public QMRDiagonalTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new DiagonalPreconditioner(A.numRows());
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTTest.java b/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTTest.java
new file mode 100644
index 0000000..5606332
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILUT;
+
+/**
+ * Test of QMR with ILUT
+ */
+public class QMRILUTTest extends QMRTest {
+
+ public QMRILUTTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILUT(new FlexCompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTest.java b/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTest.java
new file mode 100644
index 0000000..e869297
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/QMRILUTest.java
@@ -0,0 +1,41 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.CompRowMatrix;
+import no.uib.cipr.matrix.sparse.ILU;
+
+/**
+ * Test of QMR with ILU
+ */
+public class QMRILUTest extends QMRTest {
+
+ public QMRILUTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ super.createSolver();
+ M = new ILU(new CompRowMatrix(A));
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/QMRTest.java b/src/test/java/no/uib/cipr/matrix/sparse/QMRTest.java
new file mode 100644
index 0000000..3a94ea2
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/QMRTest.java
@@ -0,0 +1,39 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.sparse.QMR;
+
+/**
+ * Test of QMR
+ */
+public class QMRTest extends IterativeSolverTestAbstract {
+
+ public QMRTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createSolver() throws Exception {
+ solver = new QMR(x);
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/SPDIterativeSolverTestAbstract.java b/src/test/java/no/uib/cipr/matrix/sparse/SPDIterativeSolverTestAbstract.java
new file mode 100644
index 0000000..eed86e8
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/SPDIterativeSolverTestAbstract.java
@@ -0,0 +1,53 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.SymmDenseEVD;
+import no.uib.cipr.matrix.sparse.FlexCompRowMatrix;
+import no.uib.cipr.matrix.Utilities;
+
+/**
+ * Test of iterative solvers for SPD matrices
+ */
+public abstract class SPDIterativeSolverTestAbstract extends IterativeSolverTestAbstract {
+
+ public SPDIterativeSolverTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createMatrix() throws Exception {
+ // Create a symmetrical matrix
+ int n = Utilities.getInt(1, max);
+ int b = Utilities.getInt(Math.min(bmax, n));
+ A = new FlexCompRowMatrix(n, n);
+ Utilities.symmetryPopulate(A, b);
+
+ // Need positive eigenvalues
+ addDiagonal(A, shift);
+ SymmDenseEVD evd = SymmDenseEVD.factorize(A);
+ while (n > 0 && evd.getEigenvalues()[0] <= 0) {
+ addDiagonal(A, shift);
+ evd = SymmDenseEVD.factorize(A);
+ }
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/SparseMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/sparse/SparseMatrixTestAbstract.java
new file mode 100644
index 0000000..9f02724
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/SparseMatrixTestAbstract.java
@@ -0,0 +1,56 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.MatrixTestAbstract;
+
+/**
+ * Test of sparse matrices
+ */
+public abstract class SparseMatrixTestAbstract extends MatrixTestAbstract {
+
+ protected int bmax = 100, tmax = 10;
+
+ public SparseMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ public void testMatrixSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testTransMatrixSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testTransVectorSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testVectorSolve() {
+ // Not applicable
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/SparseStructImmutableMatrixTestAbstract.java b/src/test/java/no/uib/cipr/matrix/sparse/SparseStructImmutableMatrixTestAbstract.java
new file mode 100644
index 0000000..1a8d3ad
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/SparseStructImmutableMatrixTestAbstract.java
@@ -0,0 +1,57 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import no.uib.cipr.matrix.StructImmutableMatrixTestAbstract;
+
+/**
+ * Test of sparse matrices whose sparsity structure is immutable
+ */
+public abstract class SparseStructImmutableMatrixTestAbstract extends
+ StructImmutableMatrixTestAbstract {
+
+ protected int bmax = 100, tmax = 10;
+
+ public SparseStructImmutableMatrixTestAbstract(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ public void testMatrixSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testTransMatrixSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testTransVectorSolve() {
+ // Not applicable
+ }
+
+ @Override
+ public void testVectorSolve() {
+ // Not applicable
+ }
+
+}
diff --git a/src/test/java/no/uib/cipr/matrix/sparse/SparseVectorTest.java b/src/test/java/no/uib/cipr/matrix/sparse/SparseVectorTest.java
new file mode 100644
index 0000000..bbed048
--- /dev/null
+++ b/src/test/java/no/uib/cipr/matrix/sparse/SparseVectorTest.java
@@ -0,0 +1,100 @@
+/*
+ * Copyright (C) 2003-2006 Bjørn-Ove Heimsund
+ *
+ * This file is part of MTJ.
+ *
+ * This library is free software; you can redistribute it and/or modify it
+ * under the terms of the GNU Lesser General Public License as published by the
+ * Free Software Foundation; either version 2.1 of the License, or (at your
+ * option) any later version.
+ *
+ * This library is distributed in the hope that it will be useful, but WITHOUT
+ * ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or
+ * FITNESS FOR A PARTICULAR PURPOSE. See the GNU Lesser General Public License
+ * for more details.
+ *
+ * You should have received a copy of the GNU Lesser General Public License
+ * along with this library; if not, write to the Free Software Foundation,
+ * Inc., 59 Temple Place, Suite 330, Boston, MA 02111-1307 USA
+ */
+
+package no.uib.cipr.matrix.sparse;
+
+import java.util.Arrays;
+import java.util.Iterator;
+
+import no.uib.cipr.matrix.DenseVector;
+import no.uib.cipr.matrix.Vector;
+import no.uib.cipr.matrix.Utilities;
+import no.uib.cipr.matrix.VectorEntry;
+import no.uib.cipr.matrix.VectorTestAbstract;
+
+/**
+ * Test of SparseVector
+ */
+public class SparseVectorTest extends VectorTestAbstract {
+
+ public SparseVectorTest(String arg0) {
+ super(arg0);
+ }
+
+ @Override
+ protected void createPrimary() throws Exception {
+ int n = Utilities.getInt(1, max);
+ int m = Math.min(Utilities.getInt(max), n);
+ x = new SparseVector(n);
+ xd = Utilities.populate(x, m);
+ }
+
+ public void testSparseVectorIndices() {
+ /*
+ * MTJ subtlety in getIndex() for SparseVector. before calling
+ * getIndex(), you must call compact()... implementations may choose to
+ * do nothing in this call, but the Intel extended LAPACK
+ * implementations (and MTJ's SparseVector) require it. An alternative
+ * to vector.getIndex() is VectorMethods.getIndex(Vector) which will
+ * wrap this for you. It can take an arbitrary Vector and if it can be
+ * cast to a SparseVector will compact it and use its getIndex() method
+ * instead. (just so you're aware of this). Sam.
+ */
+
+ // check that "infinite dimensions" doesn't use infinite memory
+ SparseVector vector = new SparseVector(Integer.MAX_VALUE);
+ int[] index = vector.getIndex();
+ assert index != null;
+ assert index.length == 0;
+
+ // check that creating with double[] with zeros works
+ double[] entries = new double[5];
+ entries[0] = 0.0;
+ entries[1] = 0.0;
+ entries[2] = 1.0;
+ entries[3] = 0.0;
+ entries[4] = 2.0;
+ Vector dense = new DenseVector(entries, false);
+ vector = new SparseVector(dense);
+
+ // NOTE: must compact before calling getIndex()!!!
+ // vector.compact();
+ index = vector.getIndex();
+ assert index != null;
+ assert index.length == 5 : "expected length of 5, but got "
+ + index.length + ", with elements " + Arrays.toString(index);
+ }
+
+ public void testBug27() {
+ double[] tfVector = {0.0,0.5,0.0,0.4,0.0};
+ DenseVector dense= new DenseVector(tfVector, false);
+ SparseVector vectorTF = new SparseVector(dense);
+ vectorTF.compact();
+
+ assertTrue(vectorTF.getUsed() == 2); // vectorTF.getUsed() returns 5
+
+ for (Iterator<VectorEntry> it = vectorTF.iterator();it.hasNext();) {
+ VectorEntry ve= it.next();
+ int index = ve.index();
+ double value = ve.get();
+ assertTrue(tfVector[index]== value);
+ }
+ }
+}
--
Alioth's /usr/local/bin/git-commit-notice on /srv/git.debian.org/git/pkg-java/libmtj-java.git
More information about the pkg-java-commits
mailing list